Figure 2: Co-Detection by indEXing (CODEX) multiplex imaging workflow.
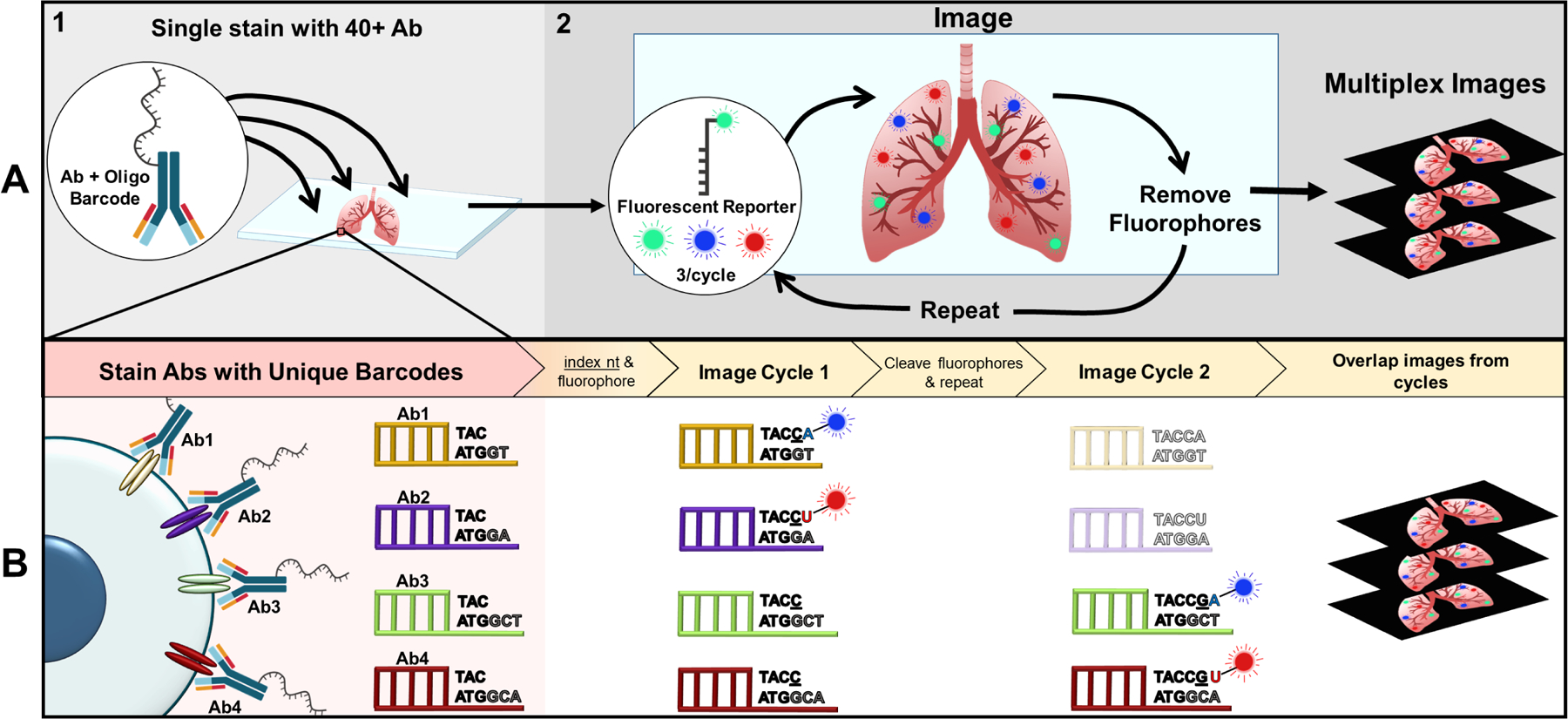
(A) Top left moving right depicting a macro-view of CODEX workflow. (A1) Tissue section (either FFPE or fresh frozen) is simultaneously stained with the entire panel of antibodies tagged with their own unique oligonucleotide barcode by the user. Of note, the commercial system currently provides ~40 barcodes that can be conjugated to any antibody of interest. (A2) The stained section then undergoes several imaging cycles automated within the fluidics device where three fluorophores are imaged and subsequently removed via TCEP cleavage each cycle (14 cycles for 40 antigens). Images are then overlapped to create a ~40 protein multiplexed image from a single tissue section. (B) Bottom depicting cellular-level view of CODEX workflow with two fluorophores (for simplicity) and indexing technology. Each antibody is conjugated to a unique barcode with a 5’ overhang. The indexing nucleotide (denoted by underlining) and two nucleotides bound to fluorophores are added to the section. As shown in cycle 1, the indexing nucleotide is incorporating into all of the barcodes, while the fluorescent nucleotides are only able to bind to their respective pre-designated barcodes to image the antibodies denoted for that cycle. Fluorophores are removed to repeat the process. Barcodes for antibodies in earlier cycles will be shorter than later cycle oligo-tags. After the antigen of interest for that cycle is imaged, the oligonucleotide will no longer be indexed, as denoted by the lighter shading in cycle 2 [70].
Abbreviations: Antibody (Ab); nucleotide (nt); FFPE (Formalin Fixed Paraffin Embedded); Tris(2-carboxyethyl)phosphine hydrochloride (TCEP)
