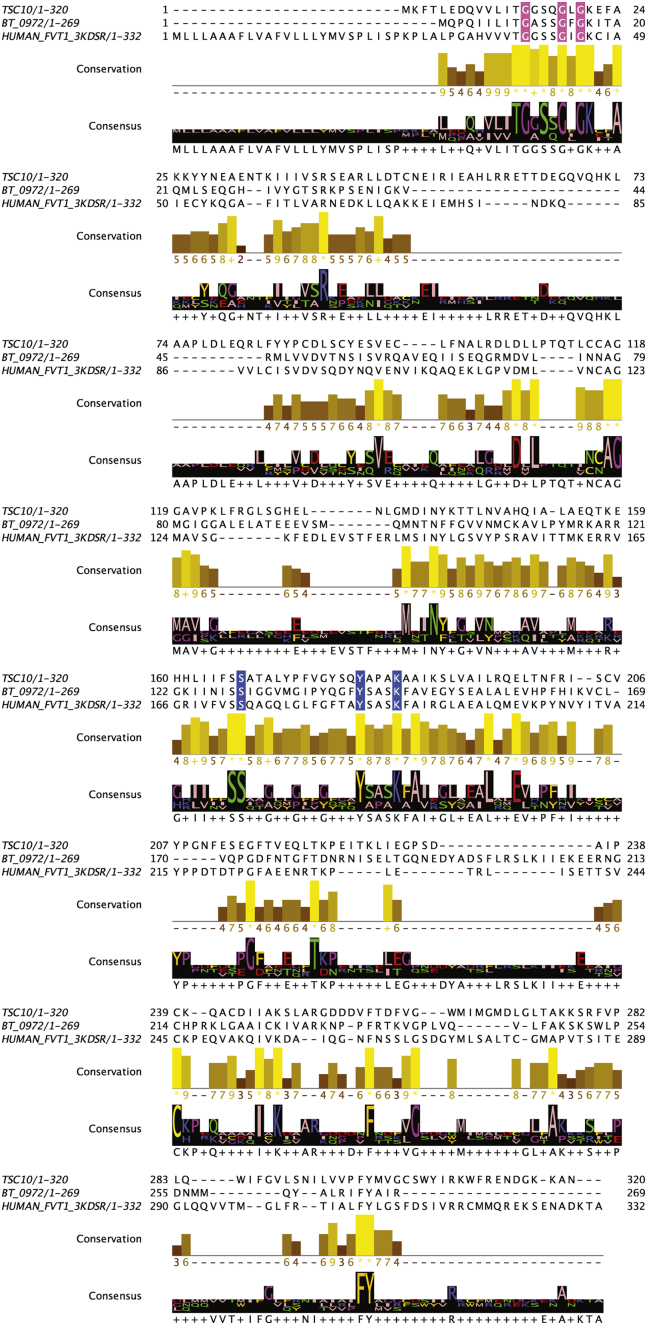
Fig. 1.
Amino acid sequence comparison of yeast, mammalian, and bacterial KDSR. Amino acid sequence alignment of Saccharomyces cerevisiae TSC10, BT_0972, and Homo sapiens FVT1 was generated using Jalview software. The putative active site motif, Tyr-X-X-X-Lys (blue) conserved in the short-chain dehydrogenase/reductase (SDR) family and the coenzyme NAD(H) or NADP(H) binding segment Gly-X-X-X-Gly-X-Gly (pink) are indicated. KDSR, ketosphinganine reductase.