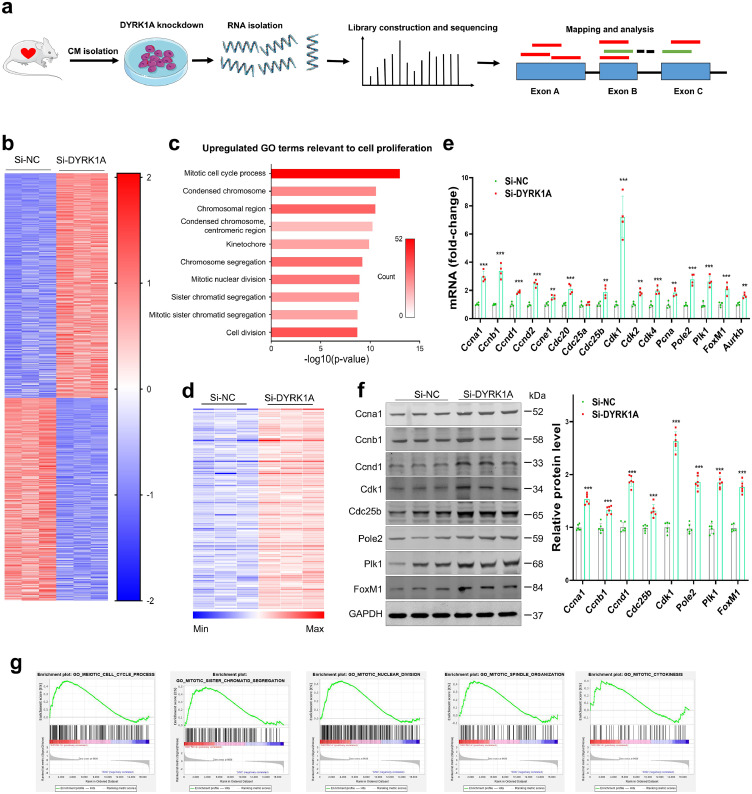
Figure 2.
DYRK1A knockdown induces transcriptional reprogramming of networks that govern cardiomyocyte cell cycle activation. (a) Schematic workflow of transcriptome analysis. (b) Heatmap profile of RNA-sequencing (RNA-seq) analysis to show differentially expressed genes in primary neonatal cardiomyocytes transfected with scramble (si-NC) or DYRK1A siRNA (si-DYRK1A) (n=3 samples per group). (c) Gene ontology (GO) analysis of upregulated genes in si-DYRK1A cardiomyocytes (relative to si-NC group) show multiple, significantly enriched GO terms relevant to cell proliferation. (d) Heatmap profile of upregulated cell cycle regulators (relative to si-NC group). (e–f) Validation of selected cell cycle regulators from RNA-seq by qRT-PCR and immunoblotting (n=4 samples per group in e; n=6 samples per group in f). Representative immunoblots (left) and quantification of protein levels (right) are shown in f. (⁎⁎P<0.01, ⁎⁎⁎P<0.001) versus si-NC group. (g) The dynamic bias of differentially expressed genes in si-NC and si-DYRK1A cardiomyocytes. Gene set enrichment analysis was used to analyze clusters of genes that regulate cell cycle. For e–f, all bars express mean ± SD and data are analyzed using the two-tailed unpaired Student's t test.