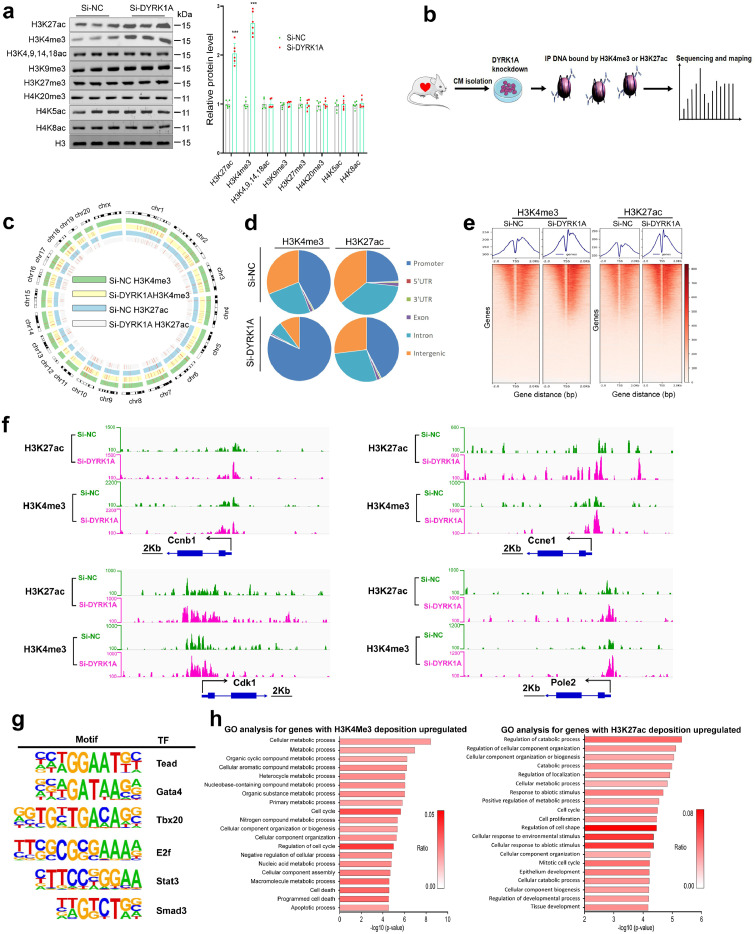
Figure 3.
DYRK1A knockdown increases deposition of H3K27ac and H3K4me3 on promoters of cell cycle genes in cardiomyocytes. (a) Changes of histone modifications induced by DYRK1A knockdown in vitro. Immunoblotting was performed to show global changes of histone modifications, using proteins extracted from cardiomyocytes in primary culture transfected with scramble (si-NC) or DYRK1A siRNA (si-DYRK1A) for 48 h (n=6 samples per group). Representative immunoblots (left) and quantification of protein level (right) are shown. ⁎⁎⁎P<0.001 versus si-NC group. (b) Schematic workflow of H3K4me3 and H3K27ac ChIP-seq. (c) Distribution of H3K4me3 or H3K27ac ChIP-seq peaks in chromosomes. The positions of H3K4me3 and H3K27ac enrichment were aligned, according to chromosome position in the outermost circle. (d) Pie chart of the genomic distribution of H3K4me3 or H3K27ac binding peaks, including promoters, 5′ UTR, 3′ UTR, exons, introns, and intergenic regions. (e) ChIP-seq density heatmaps in cardiomyocytes transfected with si-NC or si-DYRK1A. Signals were ranked by H3K4me3 and H3K27ac read intensity within ±2k bp of peak from the transcription start sites. (f) Visualization of H3K4me3 or H3K27ac ChIP-seq data tracks. Cell cycle genes were selected, and integrative genomics viewer screen shots were used to show representative peaks. (g) Motif analysis showed binding motifs for representative transcriptional factors functioning in activating cell cycle activity and cardiomyocyte proliferation. (h) GO analysis for genes with increased H3K4me3 (left) and H3K27ac (right) deposition on promoter in si-DYRK1A-treated cardiomyocytes. For a, all bars express mean ± SD and data are analyzed using two-tailed unpaired Student's t test.