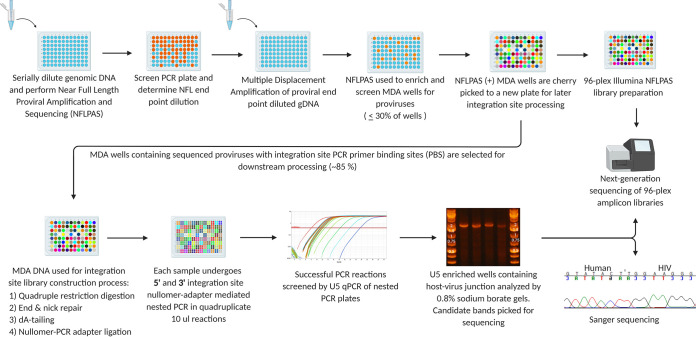
FIG 1.
Workflow for amplifying and sequencing individual HIV-1 proviruses and integration sites. gDNA extracted from cells is serially diluted and used for nested near-full-length (NFL) proviral amplification and sequencing (NFLPAS) to determine the proviral endpoint dilution factor. gDNA is then diluted and used in MDA, followed by the screening of MDA reactions for HIV-1 proviruses using NFL proviral amplification and sequencing. Gel red nucleic acid stain is used to identify NFL-positive wells. NFL-positive MDA reactions are then sequenced using the Illumina MiSeq platform, and MDA reactions containing proviruses without integration site PCR primer-binding-site deletions are selected for integration site amplification. A portion of the remaining MDA DNA is restriction digested, end repaired, dA -tailed, nullomer linker ligated, and followed by quadruplicate integration site nested PCR. PCR reactions are screened for U5 enrichment by EvaGreen qPCR, analyzed by 0.8% sodium borate agarose gel electrophoresis, and sequenced with either the Sanger or Illumina MiSeq platforms.