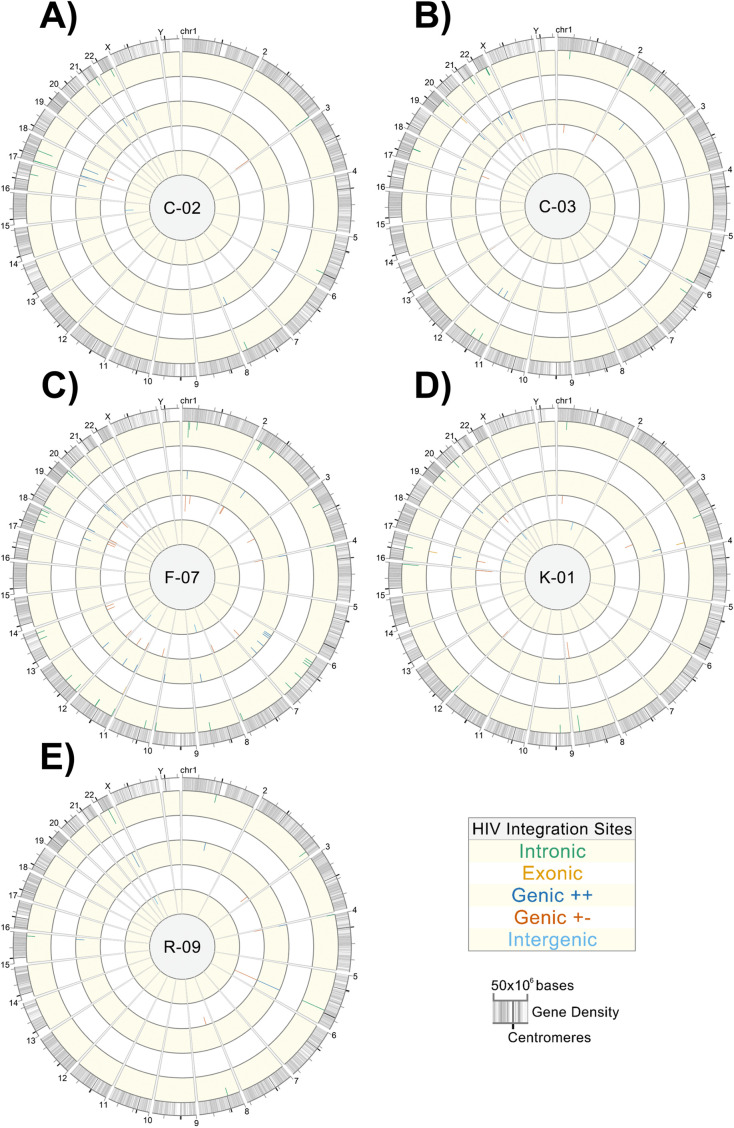
FIG 10.
Donor-specific integration site Circos plots. Sequencing reads containing the 5′-host-virus junction were used to extract integration sites from sequencing FASTQ files for alignment against the hg38 consensus genome using the UCSC BLAT tool (37). Circos plots were generated using Vgas software (38). (A) Donor C-02; (B) donor C-03; (C) donor F-07; (D) donor K-01; (E) donor R-09. Each plot depicts all amplified and sequenced proviruses and linked integration sites within a specific donor, represented by tick marks along the rings. Rings from outermost to innermost show proviruses integrated into introns, exons, genic locations in the same orientation as that of the gene, genic locations in the opposite orientation as that of the gene, and intergenic regions, respectively. The lengths of the tick marks are proportional to the number of times that an identical integration site for a given provirus was independently sequenced.