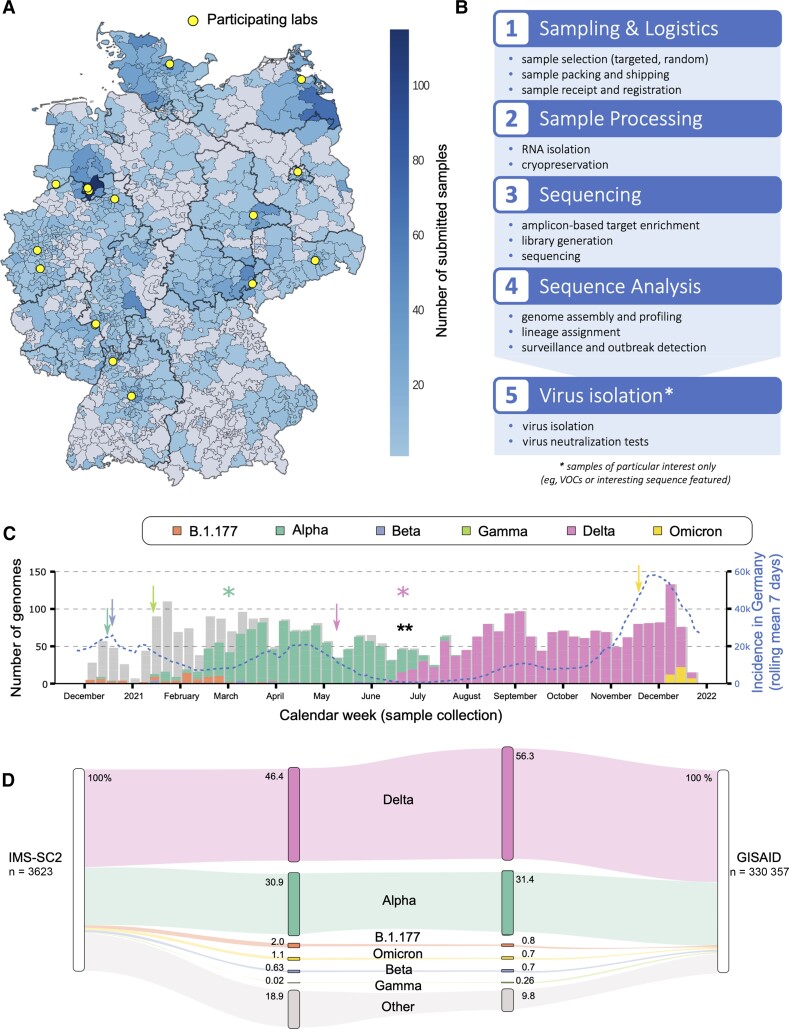
Figure 1.
A, Map visualizing the distribution and amount of submitted samples to the Integrated Molecular Surveillance for SARS-CoV-2 (IMS-SC2) project in Germany based on 3-digit zip codes. The locations of participating laboratories are highlighted in yellow. The map is based on random samples (n = 3282), suspected samples (n = 194), and unknown samples (n = 147) of which 3.42% (n = 124) were excluded due to missing or incorrect geographical data. B, IMS-SC2 workflow. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)–positive samples from multiple locations are transported to the Robert Koch Institute wet laboratory where they undergo processing for whole-genome sequencing. Sequence data are analyzed using standardized pipelines, which are updated at regular intervals to adjust for changing wet-lab protocols as well as sequence variation and evolution. All IMS-SC2 samples are cryopreserved, enabling isolation and further in vitro evaluation (eg, via neutralization assays) of SARS-CoV-2 strains displaying sequence features of interest, for example, amino acid substitutions in the spike gene that may be associated with immune evasion. C, SARS-CoV-2 lineage counts over time as captured by the IMS-SC2 network. Variants of concern (VOCs) shown are Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529). Lineage B.1.177 is also shown as an early variant that emerged in Europe in early summer 2020 [41]. Information on sublineages (such as AY sublineages for Delta) are summarized in their parent lineage. Shadow bars indicate total sequence numbers determined. Blue line displays national SARS-CoV-2 incidences (right y-axis) at the corresponding time points. Arrows denote the time points that each VOC was declared as such by the World Health Organization (Alpha: mint; Beta: blue; Gamma: light green; Delta: pink; Omicron: yellow); a black double-asterisk shows the time point at which the weekly count of Delta genomes increased significantly (Fisher exact test, P <.01); colored asterisks denote the time points when Alpha and Delta were declared predominant variants in Germany, based on sequencing data, registered case counts, and polymerase chain reaction genotyping efforts. Additional information is shown in Supplementary Table 1. D, SARS-CoV-2 lineage and corresponding sublineage proportions as captured by the IMS-SC2, compared to German genome proportions in GISAID in the same time frame. VOCs shown here are Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529), including their corresponding sublineages. Lineage B.1.177 is also shown as an early variant that emerged in Europe in early summer 2020. All remaining SARS-CoV-2 lineages are pooled into “Other.”