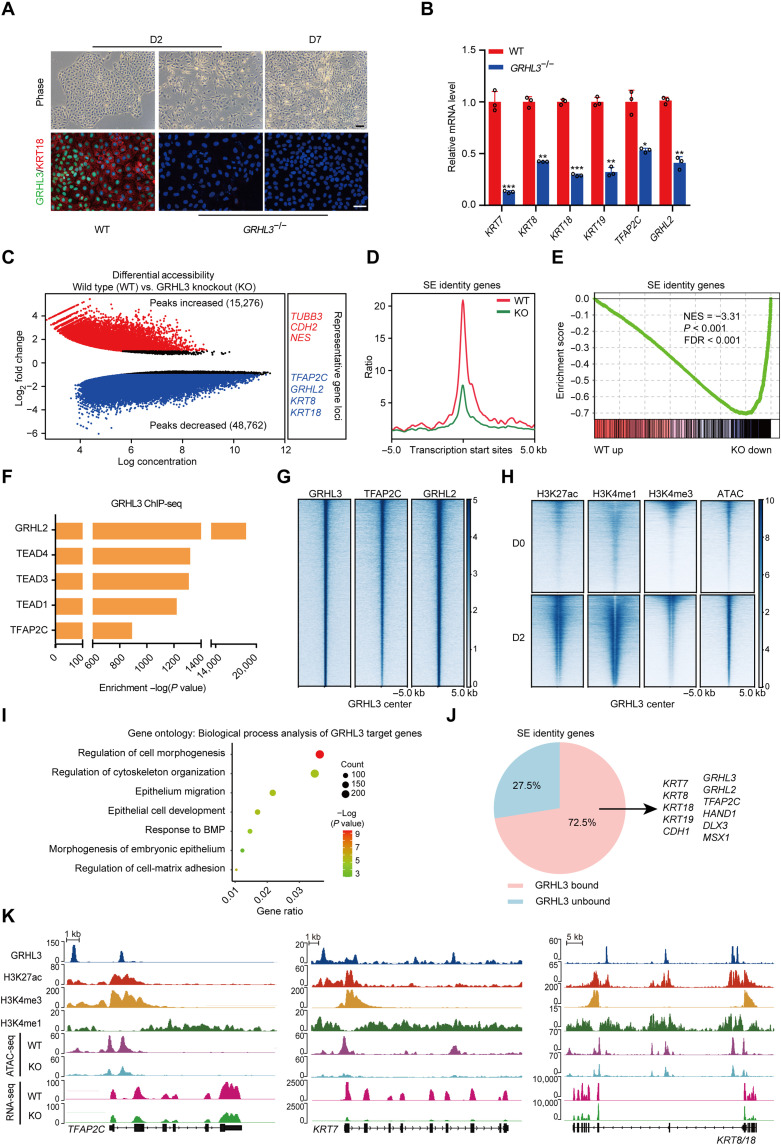
Fig. 3. GRHL3 is required to activate SE identity genes through opening their chromatin accessibilities.
(A) Wild-type (WT) and GRHL3-knockout hESCs during SE differentiation. Top: Phase contrast images. Bottom: Immunofluorescence staining of GRHL3 (green) and KRT18 (red). Scale bars, 100 μm. (B) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of representative genes in wild-type and GRHL3-knockout hESCs after 2 days of differentiation. qRT-PCR values were normalized to the values in wild-type group. Values are presented as means ± SD (n = 3 biological replicates; *P < 0.05; **P < 0.01; ***P < 0.001; t test). (C) Scatterplot of differential accessibility in wild-type versus GRHL3-knockout (KO) hESCs after 2 days of differentiation. Sites identified as significantly differentially bound [log2 fold change > 1 or < −1 and false discovery rate (FDR) < 0.05] are shown in color (red, peaks increased; blue, peaks decreased). (D) Metaplots of average ATAC-seq density around the SE identity genes in wild-type and GRHL3-knockout hESCs after 2 days of differentiation. (E) Gene set enrichment analysis of the SE identity gene set in the gene expression matrix of wild-type and GRHL3-knockout hESCs after 2 days of differentiation. NES, normalized enrichment score. (F) Enrichment of transcription factor motifs identified by HOMER at GRHL3 peaks. (G) Heatmaps of the binding signals of TFAP2C and GRHL2 at the center of GRHL3 peaks. (H) Heatmaps of H3K27ac, H3K4me1, H3K4me3, and ATAC-seq signals at GRHL3 peaks in hESCs and SE-initiating cells. (I) Gene ontology (biological process) analysis for the GRHL3 putative target genes. The GRHL3 peaks were annotated as follows: The intergenic peaks were assigned to the closest genes, and the intragenic peaks were assigned to those genes. (J) Pie chart of the percentages of SE identity genes with or without GRHL3 binding in SE-initiating cells. (K) Genome browser view of H3K27ac, H3K4me3, H3K4me1, GRHL3, ATAC-seq, and RNA-seq signal at TFAP2C, KRT7, and KRT8/18 loci.