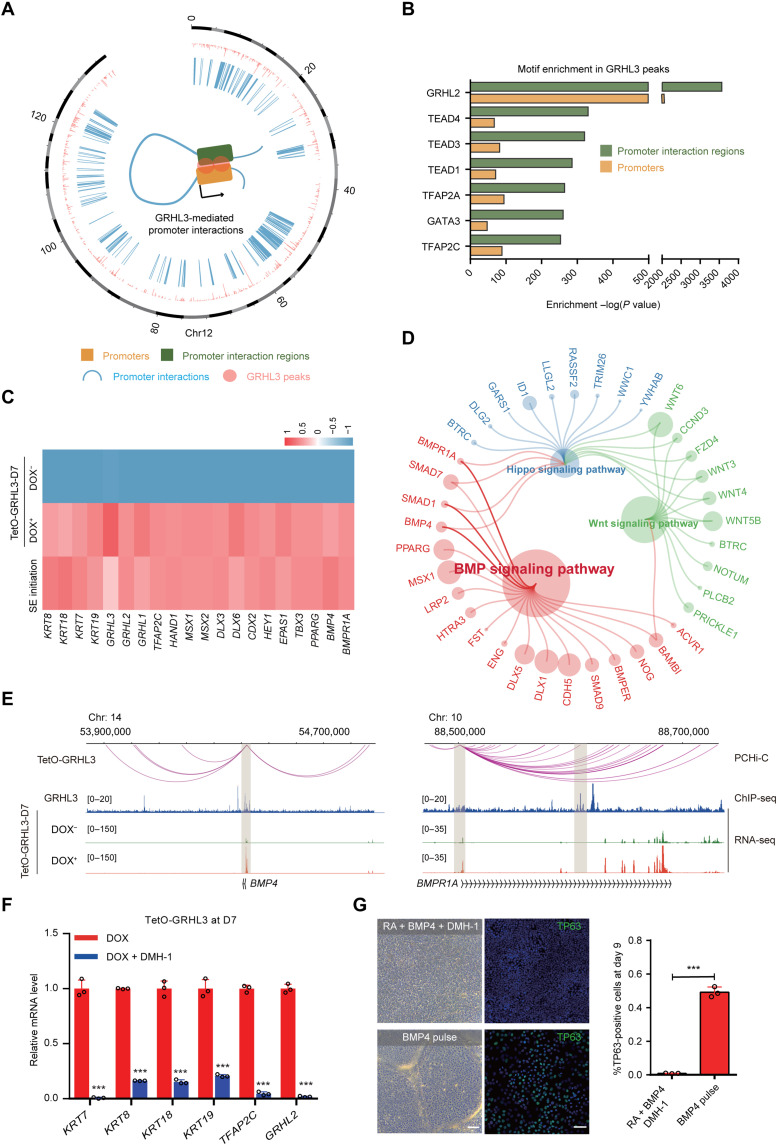
Fig. 5. Positive feedback of the BMP4-GRHL3 axis mediated SE commitment.
(A) Circos diagram of genomic promoter associated interactions mediated by GRHL3 in TetO-GRHL3+ cells. The pink color refers to GRHL3 peak. The blue color refers to promoter interactions. (B) Enrichment of transcription factor motifs identified by HOMER in GRHL3 peaks of promoter interaction regions or promoters. (C) Heatmap showing expression levels of representative genes in SE-initiating cells, TetO-GRHL3+ and TetO-GRHL3− cells. (D) Pathway enrichment analysis of representative genes in TetO-GRHL3+ cells. (E) Genome browser view of promoter interactions, GRHL3, and RNA-seq signals at BMP4 and BMPR1A loci. (F) qRT-PCR analysis of representative genes in TetO-GRHL3+ cells with or without DMH-1 for 7 days. qRT-PCR values were normalized to the values in control cells. Values are shown as means ± SD (n = 3 biological replicates; ***P < 0.001; t test). (G) Phase contrast images and TP63 staining of the differentiated hESCs after 9 days of culture. Top: hESCs treated with RA/BMP4/DMH-1. Bottom: hESC treated with a 1-day pulse of BMP4. Right: Quantification of the percentage of TP63+ cells. Values are shown as means ± SD (n = 3 biological replicates; ***P < 0.001; t test). Scale bars, 100 μm.