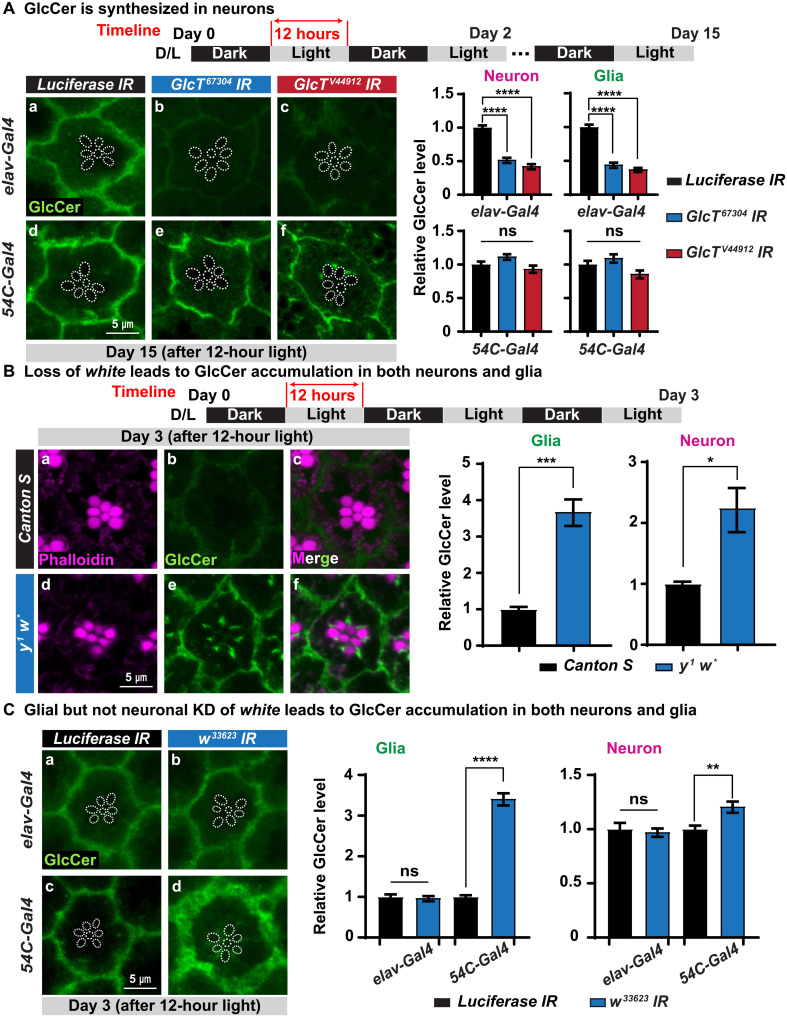
Fig. 5. Loss of white in glia causes GlcCer accumulation.
(A) GlcCer is produced in neurons. Immunofluorescent images of fly ommatidia of the indicated genotypes after 15 days of D/L cycles. Glucosylceramide synthase (GlcT) was knocked down using two different RNAi constructs in neurons driven by elav-Gal4 (top) and in glia driven by 54C-Gal4 (bottom), respectively. Neuronal but not glial knockdown of GlcT leads to a reduction of GlcCer in both neurons and glia. Dashed circles outline rhabdomeres. Relative GlcCer levels are quantified on the right. Error bars represent SEM (n ≥ 11), ****P < 0.0001. Flies that are tested in this experiment are red eyed because of the transgene. (B) Loss of white causes GlcCer accumulation. Immunofluorescent images of fly retina of the indicated genotypes after 3 days of D/L cycles. GlcCer accumulates in the ommatidia of y w but not in Canton S flies. Relative GlcCer levels are shown on the right. Error bars represent SEM (n ≥ 7), *P < 0.05 and ***P < 0.001. (C) Glial but not neuronal knockdown (KD) of white causes GlcCer accumulation. Immunofluorescent images of fly retina of the indicated genotypes after 3 days of D/L cycles. The white mRNA was knocked down using a UAS-RNAi construct expressed in neurons by elav-Gal4 (top) and in glia driven by 54C-Gal4 (bottom), respectively. Knockdown of white in glia leads to an accumulation of GlcCer in both neurons and glia, whereas neuronal knockdown in neurons does not cause a phenotype. Relative GlcCer levels are quantified on the right. Error bars represent SEM (n = 11), **P < 0.01 and ****P < 0.0001. Flies that are tested in this experiment are red eyed because of the transgenes.