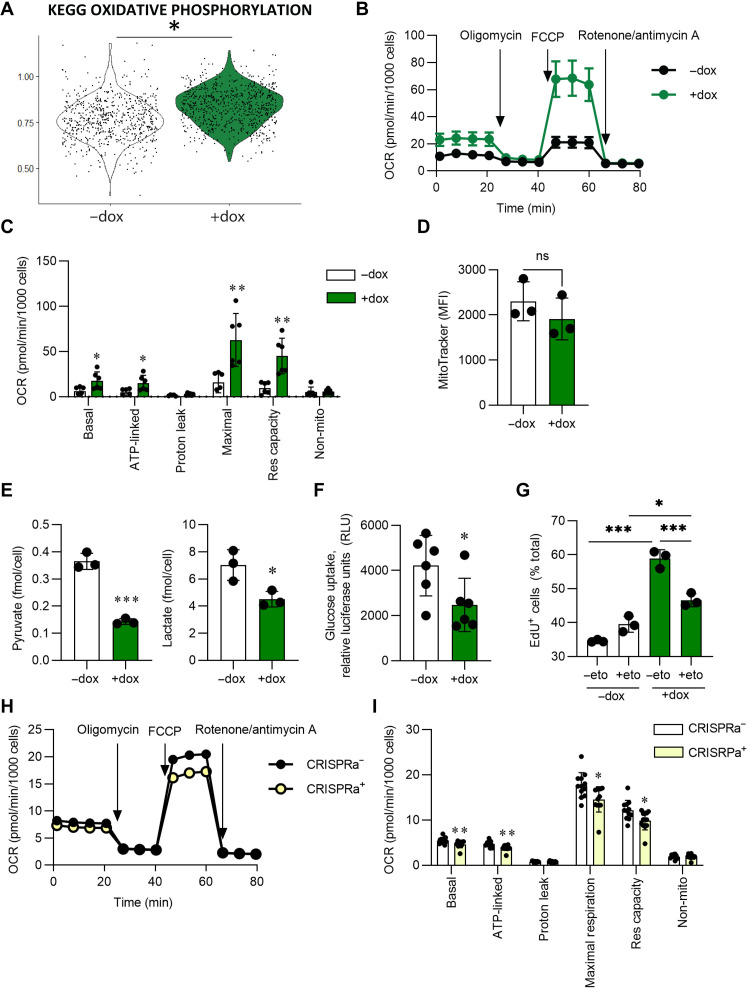
Fig. 6. DSP expression influences mitochondrial respiration.
CRISPRi-iAT2s were transduced with DSP gRNA and treated without (−dox) or with (+dox) doxycycline (A to G). (A) Module score for KEGG oxidative phosphorylation of scRNA-seq. (B and C) Basal oxygen consumption rate (OCR) was measured, followed by injection of oligomycin, FCCP, and rotenone + antimycin A, as indicated. Data were normalized by cell count after the assay was complete. n = 6 technical replicates of a differentiation. (D) MitoTracker staining was quantified by flow cytometry, represented by MFI; n = 3 experimental replicates of independent wells of a differentiation. (E) Pyruvate or lactate was measured in the supernatant and normalized by cell count; n = 3 experimental replicates of independent wells of a differentiation. (F) Glucose uptake was determined using 2-DG and measuring luciferase; n = 3 experimental replicates of independent wells of two differentiations. (G) Cells were treated with 10 μM etomoxir or vehicle for 21 days. One day before harvest, cells were incubated with EdU. EdU incorporation was measured by flow cytometry. n = 3 experimental replicates of independent wells of a differentiation. (H and I) iAT2s were transduced with DSP gRNA ± CRISPRa lentivirus. Basal OCR was measured, followed by injection of oligomycin, FCCP, and rotenone + antimycin A, as indicated. Data were normalized by cell count after the assay was complete. n = 10 technical replicates of a differentiation. All error bars represent SD. Statistical significance was determined using unpaired, two-tailed Student’s t test or a one-way ANOVA with a Tukey multiple comparison test; *P < 0.05, **P < 0.005, and ***P < 0.001.