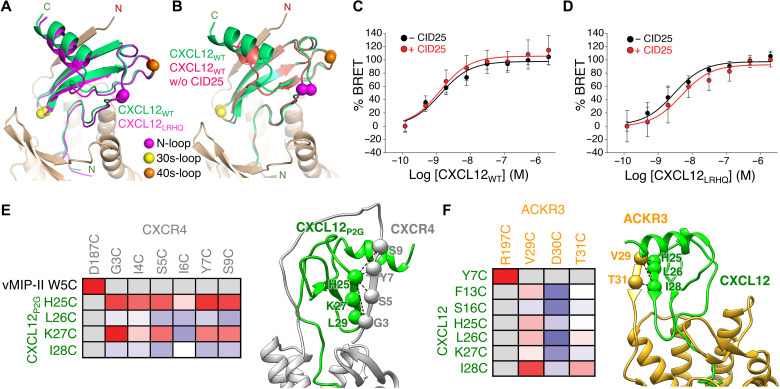
Fig. 3. The binding mode of CXCL12 on ACKR3 is not altered by CID25 and is distinct from that of CXCR4.
(A) Comparison of CID25-ACKR3·CXCL12WT-CID24 (PDB entry 7SK3) with CID25-ACKR3·CXCL12LRHQ-CID24 (PDB entry 7SK4) reveals a nearly identical mode of interaction. (B) Comparison of CID25-ACKR3·CXCL12WT-CID24 with ACKR3·CXCL12WT-CID24-NB (PDB entry 7SK5) reveals that CID25 does not greatly influence the chemokine pose. (C and D) Bioluminescence resonance energy transfer (BRET) assays of GFP10–β-arrestin 2 recruitment to ACKR3-RlucII as a function of CXCL12WT (C) or CXCL12LRHQ (D) in the absence or presence of 2 μM CID25. The extracellular Fab had an insignificant effect on the negative logarithm of the half maximal effective concentration (−8.87 and −8.89 without or with CID25 for CXCL12WT, respectively, P > 0.5; −8.53 and −8.32 without or with CID25 for CXCL12LRHQ, respectively, P > 0.3). The Emax was also not affected by CID25 for either chemokine (P > 0.05). Statistics were determined by extra sum-of-squares F test. Data are a composite of three separate experiments measured in triplicate and normalized to the −CID25 dataset for each ligand. Error bars correspond to SD across the three experiments. (E and F) Disulfide cross-linking of ACKR3 and CXCR4 with CXCL12 confirms that the two complexes have distinct chemokine binding poses. Heatmaps depicting the relative cross-linking efficiency from coexpression of single cysteine mutants of CXCL12 and ACKR3 versus CXCR4 and CXCL12P2G [a point mutant of CXCL12 that functions as an antagonist (41)] in Sf9 cells quantified by flow cytometry (table S3). The cross-link propensity is depicted from blue (absent) to red (strong). Positive disulfide cross-links are noted with spheres and dashed lines on the accompanying structures. The CXCR4 cross-linking data were adapted from (22).