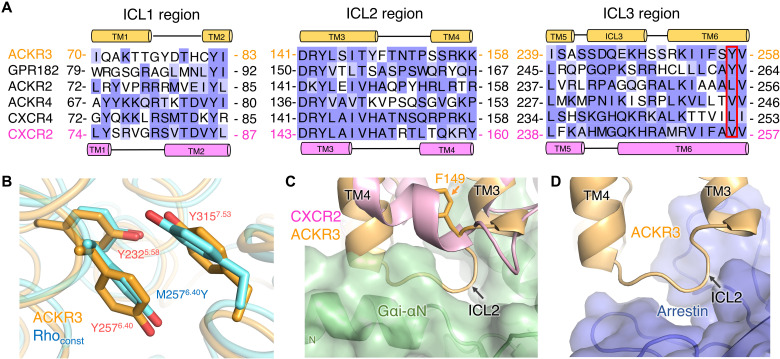
Fig. 6. Residues within the intracellular cleft of ACKR3 assume an active-like conformation, but ICL2 assumes an inactive configuration that seems incompatible with G protein coupling.
(A) Sequence alignment of ICLs in atypical chemokine receptors versus CXCR4 and CXCR2. Secondary structures are shown on top and bottom for ACKR3 and CXCR2, respectively. Positions corresponding to ACKR3-Y2576.40 are bounded with a red box. ACKR1 is omitted because of its high sequence divergence. (B) Orientation of ACKR3-Y2325.58, ACKR3-Y2576.40, and ACKR3-Y3157.53 (PDB entry 7SK3, orange) compared with the N2C/M2576.40Y/D282C constitutively active rhodopsin mutant (Rhoconst; PDB entry 4A4M, cyan). (C) Superposition of ACKR3 (PDB entry 7SK3, orange) with CXCR2 in complex with CXCL8 and GαiGβ1γ2 (PDB:6LFO, CXCR2 in pink, Gαi in green). The nonconserved side chain of F149 in ICL2 of ACKR3 [orange sticks, see (A)] may prevent the kink that forms in TM4 of CXCR2. (D) Superposition of ACKR3 (PDB entry 7SK3, orange) with the M2 muscarinic receptor–arrestin complex (PDB:6U1N; only arrestin is shown in blue).