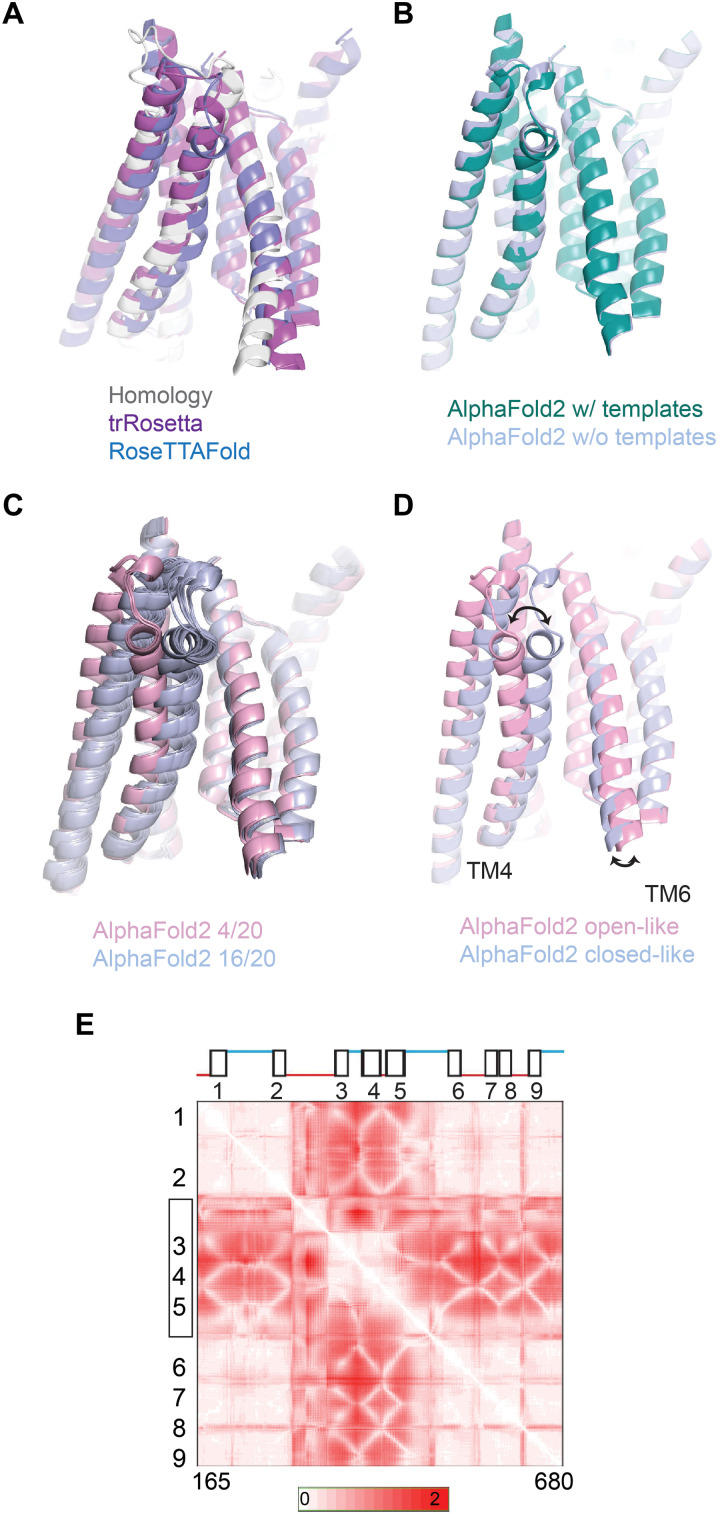
Fig. 1. Similarity in TMC1 structural predictions.
(A) Structural models for mouse TMC1. Shown in foreground are TM3, TM4, TM6, and TM8 viewed from within the membrane plane. Gray represents an I-TASSER model based on homology to TMEM16 channels. Purple shows a prediction by trRosetta. Blue is a template-free prediction by RoseTTAFold. The three models are nearly identical. (B) Structural model for TMC1 from AlphaFold2 is shown in green. The template-free AlphaFold 2 model is shown in blue. In the pore region, they are the same within 1- to 2-Å root mean square deviation. (C) Multiple structural models from AlphaFold2, generated with multiple seed parameters to achieve diversity. In 20 model iterations, 16 grouped in one conformation [blue; similar to that in (B)] and 4 grouped in a different conformation (pink). (D) Representative models from the two groups of conformations in (C). The conduction pathway for ions is between TM4 and TM6. In the more open conformation (pink), TM4 is more distant from TM6 by about 5 Å at the constricted region of the pore. (E) SD maps for all residue-residue distances for residues 165 to 680. For each residue pair, more intense red represents a larger change in distance between the two conformations. TM helices are numbered. The N terminus, C terminus, and TM10 were not included in the analysis.