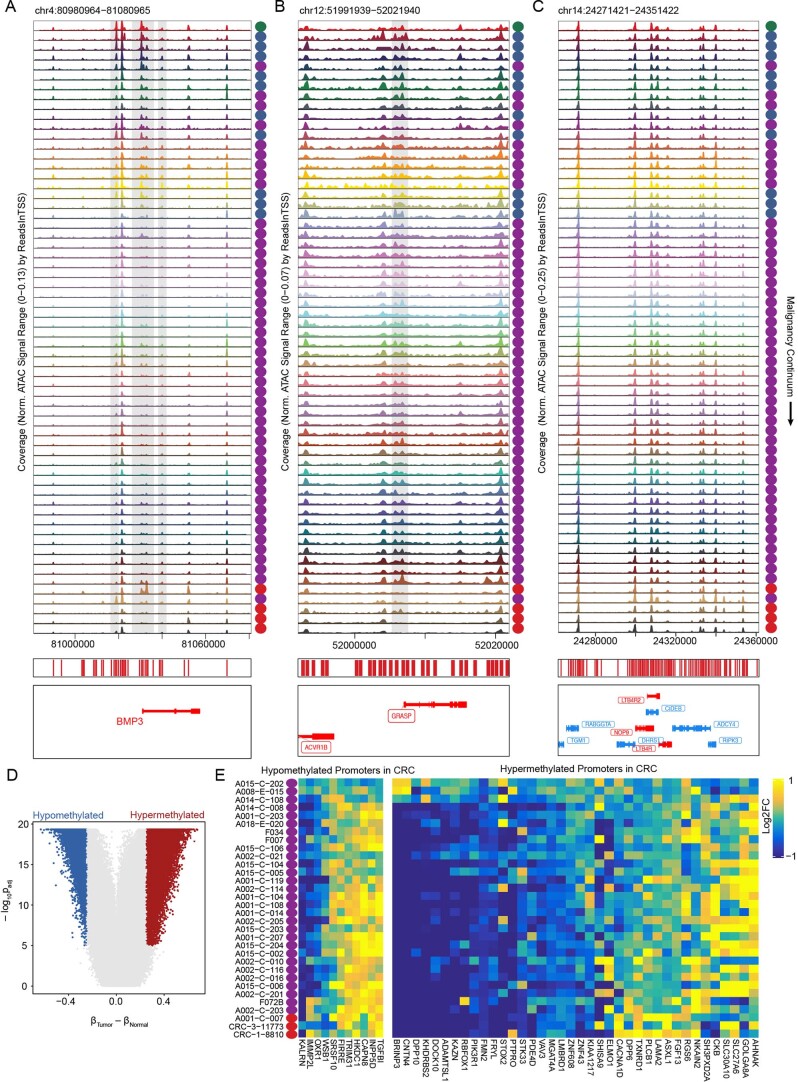
Extended Data Fig. 9. Accessibility changes in regions hypermethylated in CRC.
(a–c) Accessibility tracks around BMP3 (a), GRASP (b), and CIDEB (c), which are hypermethylated in CRC. (d) Adjusted p-value and mean difference in β-value cutoffs used to determine differentially methylated probes. P values were determined using the two-sided Wilcoxon test and were adjusted with the Benjamini-Hochberg method for multiple hypothesis testing. (e) Genes that were significantly differential along the malignancy continuum that also have differentially methylated probes within 500 bp of their TSS in TCGA 450 K methylation data. Genes are grouped into a heatmap of those with hypermethylated probes in their promoters and a heatmap of those with hypomethylated probes in their promoters.