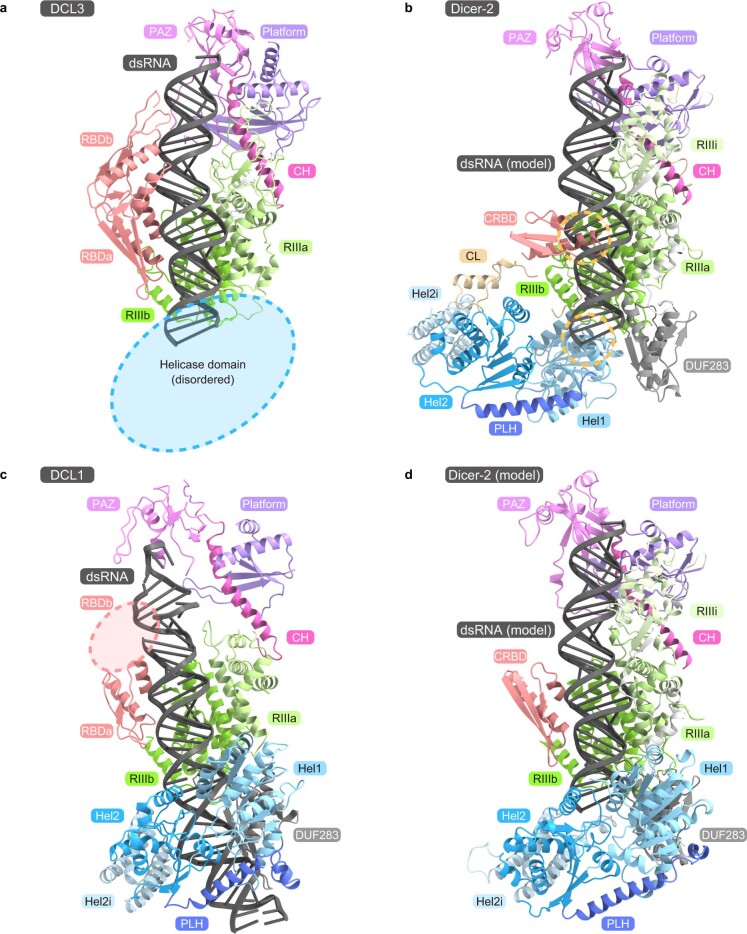
Extended Data Fig. 8. Structural comparison between Dicer-2, DCL1, and DCL3.
(a) Structure of the DCL3–dsRNA complex (PDB: 7VG3). The amino-terminal helicase domain is disordered in the structure. (b) Model of the Dicer-2–dsRNA complex. The DCL3–dsRNA structure was superimposed onto the Dicer-2 structure, based on their RNase III domains, and DCL3 was then omitted. Predicted steric clashes are indicated by dashed circles. (c) Structure of the DCL1–dsRNA (pri-miRNA) complex (PDB: 7ELD). The carboxy-terminal RBDb domain is disordered in the structure. (d) Model of the Dicer-2–dsRNA complex. The Dicer-2 structure was predicted by AlphaFold2, and the dsRNA and CRBD were modeled based on the DCL3–dsRNA structure. Unstructured regions in the RIIIb and CL domains are omitted for clarity. In the model, the helicase domain interacts with the DUF283 domain and recognizes the dsRNA substrate, as in the DCL1–dsRNA structure.