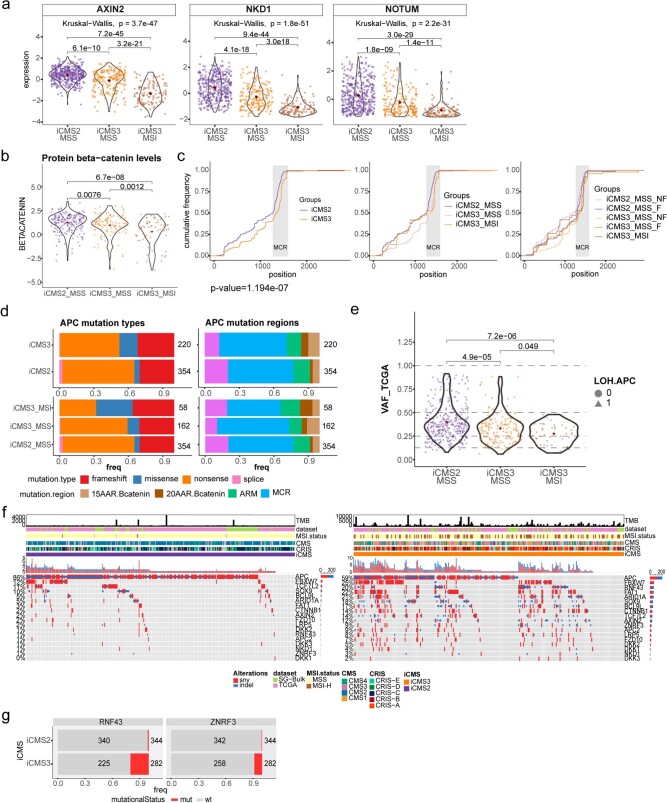
Extended Data Fig. 6. Wnt pathway alterations in iCMS.
a. Scaled gene expression of selected Wnt pathway genes in iCMS2_MSS (n = 389), iCMS3_MSS (n = 195) and iCMS3_MSI (n = 116), from TCGA and SG-Bulk datasets. b. Beta-catenin protein levels, by RPPA, in iCMS2_MSS (n = 216), iCMS3_MSS (n = 117) and iCMS3_MSI (n = 67) from TCGA data. P-values for pairwise comparisons are by two-sided Wilcoxon rank-sum test with no correction; overall p-value is by Kruskal-Wallis test. c. Cumulative frequencies of truncating (nonsense and frameshift) APC mutations by position from TCGA and SG-Bulk patients with APC mutations (n = 342), in iCMS2 and iCMS3 (left), iCMS2-MSS vs iCMS3-MSS vs iCMS3-MSI (middle), as well as IMF 5 groups (right). P-value is by two-sided Kolmogorov–Smirnov test. d. Proportion of APC mutation types (left) and regions (right) in samples with APC mutations from TCGA and SG-Bulk data (n = 574). Comparisons are between iCMS2/iCMS3 (top), and iCMS2_MSS/iCMS3_MSS/iCMS3_MSI (bottom). e. APC variant allele frequency (VAF) in iCMS2_MSS (n = 343), iCMS3_MSS (n = 172) and iCMS3_MSI (n = 46) from TCGA data. P-values are by two-sided Wilcoxon rank-sum test with no correction f. Mutational landscape of selected Wnt pathway genes in iCMS2 (left, n = 344) and iCMS3 (right, n = 281) samples in TCGA and SG-Bulk datasets. g. Proportion of samples in TCGA and SG-Bulk datasets (n = 626) with wild type (wt) or mutations (mut) in RNF43 (top) and ZNRF32 (bottom); number of samples in each group is labelled.