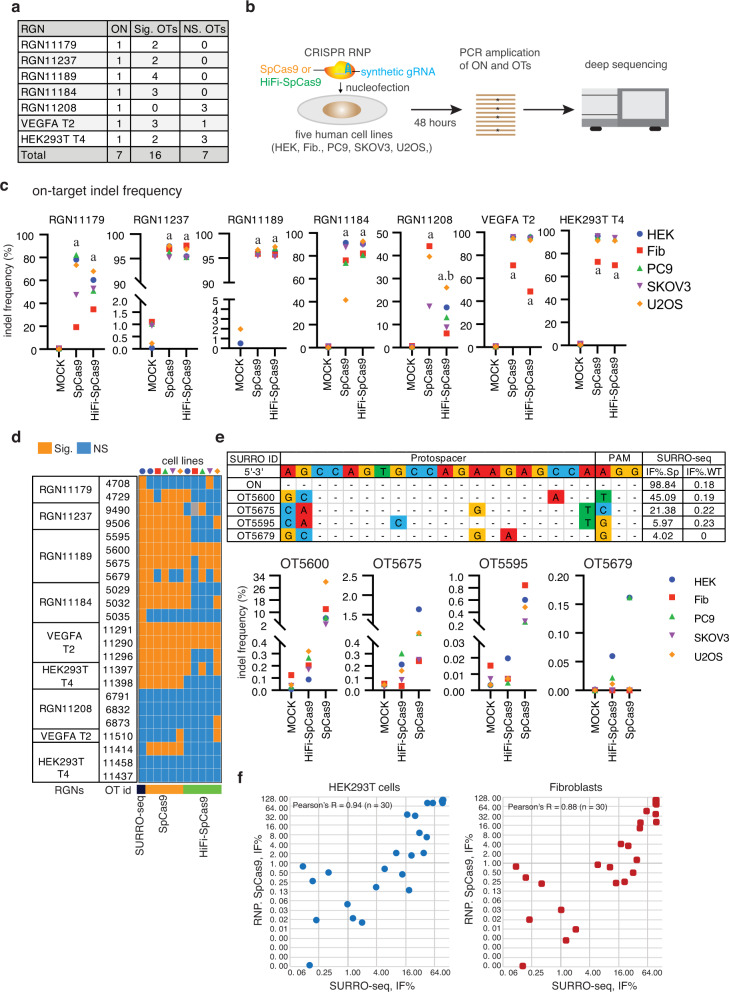
Fig. 4. Validation of endogenous OTs in five human cell lines by deep sequencing.
a Overview of RGNs, SURRO-seq identified Sig. OTs and NS. OTs selected for validation. b Schematic illustration of the experiments. RNP, ribonucleoprotein; HEK, HEK293T cell; Fib, human skin-derived fibroblasts. c Dot plot of on-target indel frequencies (indel reads/total reads %) in the CRISPR RNP edited cells. Indel frequency values were showed in Supplementary Data 4. d Heatmap summary of the RGN OTs evaluated by SURRO-seq and deep sequencing in five human cells lines. Indel frequency values were showed in Supplementary Data 4. e Example of indel frequencies of four OTs for RGN11189 measured by SURRO-seq and by deep sequencing of RGN edited human cell lines. f Scatter plots of indel frequencies for 7 on-target and 23 off-targets (referred to 4a), measured by SURRP-seq and by amplicon sequencing of the corresponding endogenous loci in RNP nucleofected cells (HEK293T and Fibroblasts). Extended plots for all cells can be found in Supplementary Data 4 (sheet 4.9).