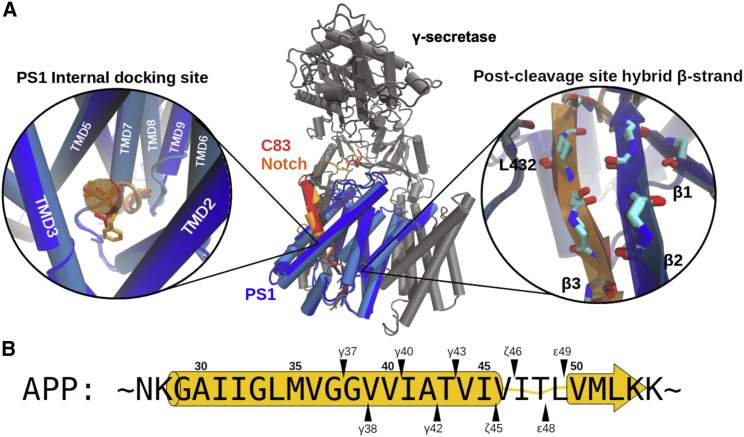
Figure 1.
Cryo-EM structure of substrate-bound γ-secretase and APP cleavage sites. (A) Superposition of the Notch1-bound (PDB: 6IDF) and C83-bound (PDB: 6IYC) γ-secretase structures resolved by cryo-EM (39,40) with γ-secretase cartoon model colored in gray and the catalytic subunit PS1 in dark blue (from PDB: 6IDF) or light blue (from PDB: 6IDF). The backbone RMSD of PS1, including L73-T291 and E376-I467, between two structures is 0.85 Å. Substrates are shown in red (C83) and orange (Notch1). (Left zoom-in) The PS1 internal docking site with V44-I45 of C83 (red) and F1748-F1749 of Notch1 (orange) aligned. (Right zoom-in) The hybrid β sheet formed downstream of the substrate cleavage site. (B) Sequence of APP and its consecutive cleavage sites targeted by γ-secretase. The two production lines are depicted above and below the APP sequence. The yellow cartoon representation in the background indicates the range of the helix-loop-strand conformation of the APP TMD in the bound state. To see this figure in color, go online.