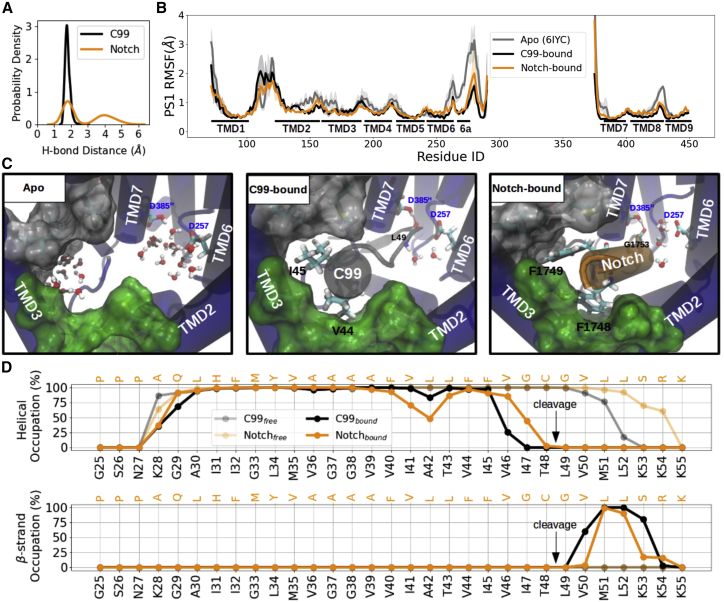
Figure 2.
MD simulations of unbound γ-secretase and bound to C99 and Notch substrates. (A) Probability density distribution of the catalytic hydrogen bond distance. (B) Residue-wise root-mean-square fluctuation (RMSF) of the γ-secretase catalytic subunit PS1. The unbound γ-secretase starting structures were used based on PDB 6IYC (gray). (C) View into the PS1 internal docking site in the unbound form (left), C99-bound (middle), and Notch-bound (right) γ-secretase complexes. PS1 is shown in blue cartoon representation, C99 in gray, and Notch in orange. The sub-pocket formed between TMD2 and TMD3 is shown as green surface, and the sub-pocket formed by TMD3-TMD5 and TMD7 is indicated as light gray surface. All residues defining these two pockets are listed in Table 2. Water molecules are shown in van der Waals (vdw) + bond representation. V44, I45, and the backbone of L49 of C99 and F1748, F1749, and the backbone of G1753 of Notch are shown in the licorice representation. The catalytic hydrogen bond is indicated as red dashed line between the substrate scissile bond and the protonated aspartic acid. (D) Secondary structure analysis of C99 (black) and Notch (orange) in γ-secretase bound form (solid line) and unbound (mostly helical) form (embedded in a membrane, transparent line). Helical and β strand occupancies are calculated as averages over the whole trajectories. To see this figure in color, go online.