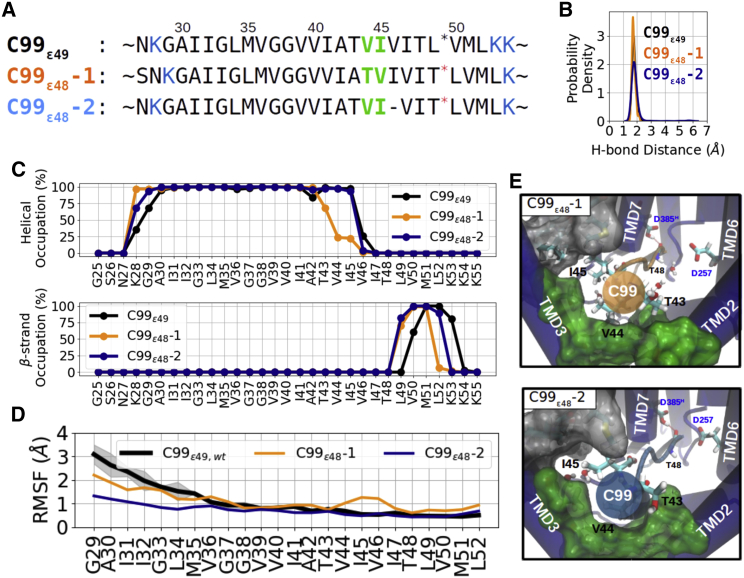
Figure 3.
MD simulations of γ-secretase in complex with C99 shifted for cleavage at ε48 site. (A) Sequence alignments of the two models termed C99ε48-1 (orange), and C99ε48-2 (dark blue) relative to the residue positions in C99ε49 (black). Membrane-anchoring residues K28, K53, and K54 are marked in blue and residues residing in the hydrophobic pockets are marked in green. (B) Sampled probability density of the catalytic hydrogen bond distance in the MD simulation. (C) Secondary structure analysis of simulation on C99ε49 (black), C99ε48-1 (orange), C99ε48-2 (dark blue) bound to γ-secretase complexes. (D) RMSF of the substrates. (E) View (from the extracellular side) into the PS1 internal docking site of the two models C99ε48-1 (upper) and C99ε48-2 (lower) (representation same as in Fig. 2C). To see this figure in color, go online.