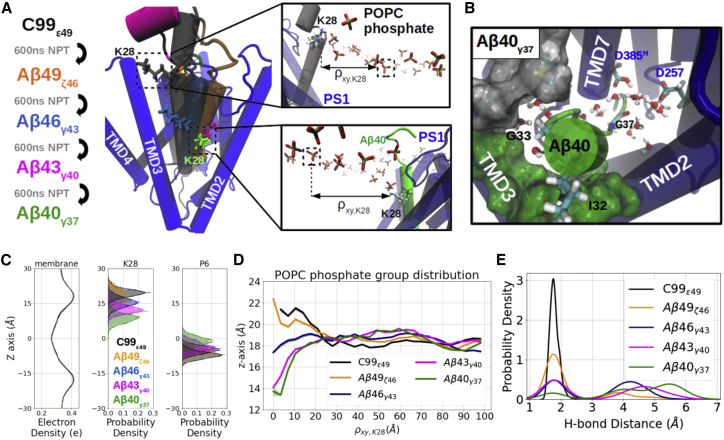
Figure 5.
Comparative modeling and simulations of Aβn-γ-secretase complexes. (A) Schematic views of the modeling workflow and the top views (from the extracellular domain) of the binding poses of C99ε49 (black), Aβ49ζ46 (orange), Aβ46γ43 (blue), Aβ43γ40 (magenta), and Aβ40γ37 (green). Membrane-anchoring residue K28 of each substrate is shown explicitly in the licorice representation. Zoom-in views of the C99ε49 (upper) and Aβ40γ37 (lower) show how the position of K28 influences the local distribution of the POPC phosphate groups with radial distance from K28 on the xy plane denoted by ρxy,K28. (B) View into the PS1 internal docking site in the Aβ40γ37-bound γ-secretase (representation same as in Fig. 2C) (C). Distribution of the membrane electron density (left), membrane-anchoring residue K28 (middle), and substrate P6 (right) along the z axis in different Aβn-γ-secretase complexes. (D) Average z axis of the POPC phosphate on the extracellular side distributed along the radial distance ρxy,K28. (E) Probability density of the catalytic hydrogen bond distance. To see this figure in color, go online.