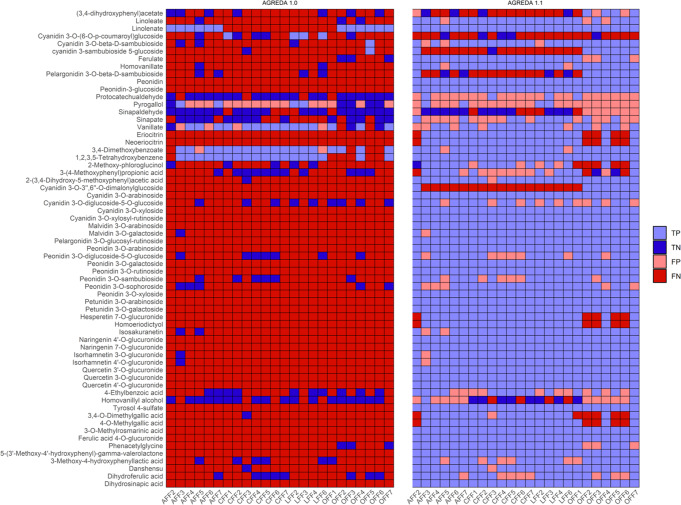
Fig. 4. Comparison between the predictions of AGREDA_1.017 and AGREDA_1.1 with in vitro experiments.
Representation of the presence of 63 output microbial compounds predicted in AGREDA_1.017 and AGREDA_1.1 to derive from the fermentation of lentils with children feces and measured with an untargeted metabolomics approach. “AFF2”, “AFF3”, “AFF4”, “AFF5”, “AFF6” and “AFF7” denote samples 2, 3, 4, 5, 6 and 7 from children allergic to cow’s milk, respectively; “CFF1”, “CFF2”, “CFF3”, “CFF4”, “CFF5”, “CFF6” and “CFF7” denote samples 1, 2, 3, 4, 5, 6 and 7 from celiac children, respectively; “LFF2”, “LFF3”, “LFF4” and “LFF6” denote samples 2, 3, 4 and 6 from lean children, respectively; “OFF1”, “OFF2”, “OFF3”, “OFF4”, “OFF5”, “OFF6” and “OFF7” denote samples 1, 2, 3, 4, 5, 6 and 7 from obese children, respectively; TP true positives, TN true negatives, FP false positives, FN false negatives. Source Data are provided as a Source Data file.