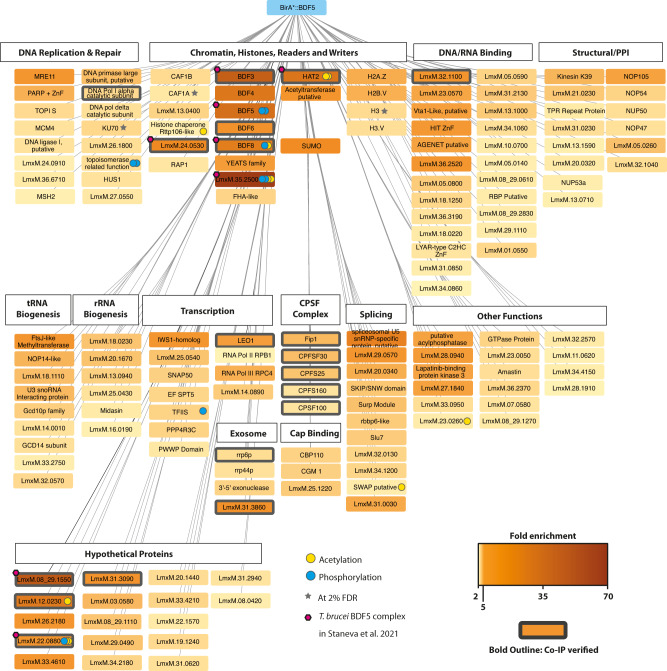
Fig. 5. The BDF5-proximal proteome determined by XL-BioID.
Network indicates proteins determined to be spatially enriched in proximity to BirA*::BDF5 following in vivo biotin labelling and DSP crosslinking during the XL-BioID workflow. Biotinylated material was enriched using streptavidin magnetic beads, trypsin digested and analysed by LC-MS/MS. SAINTq analysis was performed to determine statistically significant enrichment scores. Fold enrichment values are encoded in the colour intensity of the protein boxes; these were calculated from label free protein intensities against a control expressing CLK2::BirA*, from 3 replicate experiments. A bold outline on a box indicates that BDF5 co-purified with this protein in a reciprocal co-immunoprecipitation experiment. If a post-translational modification (PTM) was detected for a protein, this is indicated using a coloured circle. Proteins are grouped by functional annotations or previously published data of complexes in Leishmania or Trypanosoma. Proteins represented are those identified at 1% false-discovery rate (FDR), those marked with a grey star denote those identified at 2% FDR for selected proteins. CPSF stands for Cleavage and Polyadenylation Specificity Factor complex, PPI stands for protein-protein interactions. Magenta hexagons indicate members of the BDF5, BDF3, HAT2 complex reported in T. brucei.