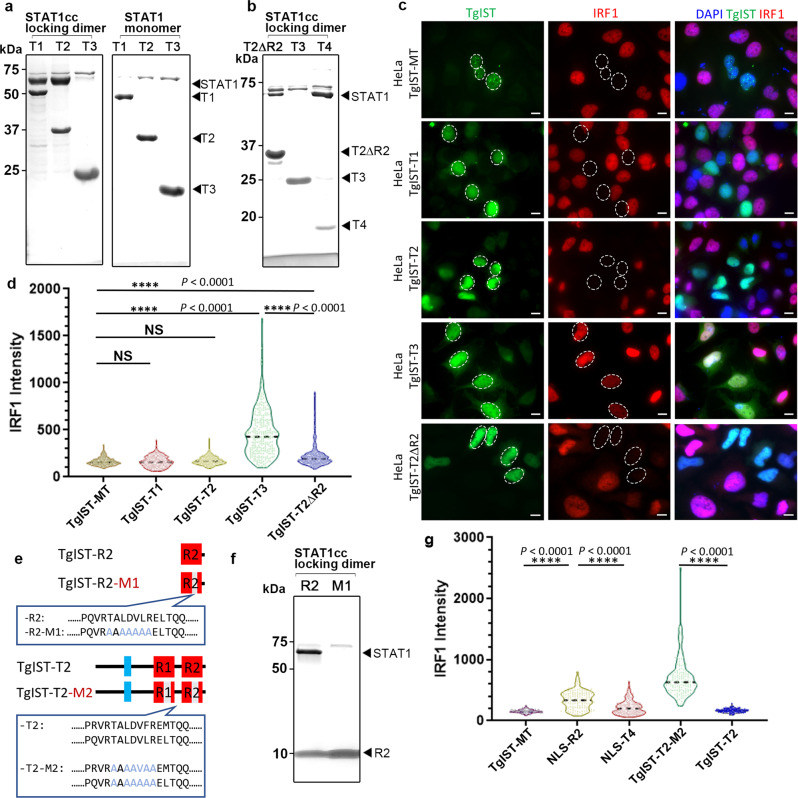
Fig. 2. The TgIST repeats are required for STAT1 dimer binding and inhibition of IFN-γ signaling in host cells.
a, b Co-purification of STAT1cc or STAT1 with TgIST constructs as defined in Fig. 1d following co-expression in E. coli. Co-purifications were performed by capturing TgIST His6-tagged constructs with nickel resin. SDS-PAGE gels stained with Coomassie blue. Representative gel images of two independent experiments with similar results are shown here. c Representative images showing induction of IRF1 in transfected Hela cells. Cells transiently expressing GFP-tagged TgIST constructs for 24 h were activated with IFN-γ for 6 h followed by staining for GFP (green), IRF1 (red) and DAPI (blue). Scale bar = 10 μm. Expression level of STAT1 remained unchanged (see Supplementary Fig. 3). Representative micrographs of two independent experiments with similar results are shown here. d Distribution of the IRF1 intensity collected from at least 250 TgIST-expressing HeLa. ****P ≤ 0.0001 with Kruskal–Wallis’s test for multiple comparisons, and with Dunn’s test for non-parametric correction. TgIST constructs correspond to those shown in (c) and are listed only with the suffix denting the construct. Data presented as violin plot and median (dotted line) from two independent experiments are shown. e Sequences of wild type TgIST-R2 containing a single repeat and a mutated version with an altered core region defined by changing TALDVLR to AAAAAAA (TgIST-R2-M1). Sequences of wild type TgIST-T2 containing two repeats and a mutated version with an altered core region defined by changing TALDVLR to AAAAAAA (TgIST-T2-M2). (f) TgIST-R2 and TgIST-R2-M1 tagged with His6 were co-expressed with the STAT1cc locked dimer. Purification of the His-tagged TgIST proteins on nickel resin resulted in co-purification of STAT1cc with the wild-type TgIST-R2 (R2) construct but not the R2-M1 mutant (M1). SDS-PAGE gel stained with Coomassie blue. Representative gel image of two independent experiments with similar results are shown here. g The core repeat region was required for blocking IFN-γ signaling as detected by induction of IRF1. IRF1 intensity levels of HeLa cells stained as above. Data were collected from at least 250 TgIST-expressing (NLS-R2 or NLS-T4: R2 or T4 protein with nuclear localization sequence) HeLa cells and expressed as violin plots. ****P ≤ 0.0001 with Kruskal–Wallis’s test for multiple comparisons, and with Dunn’s test for non-parametric correction. Data presented as median (dotted line) from two independent experiments. Source data are provided in the Source Data file.