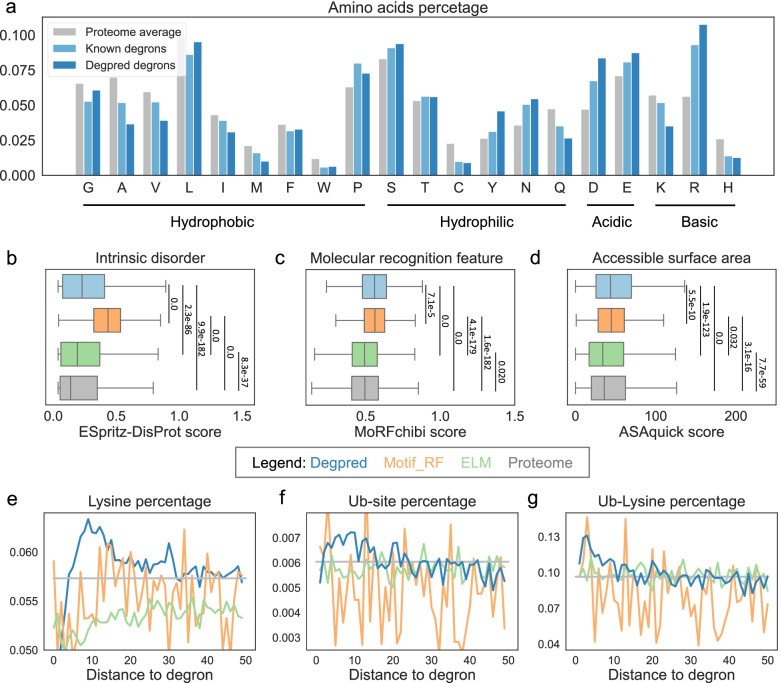
Fig. 3.
Properties of Degpred degrons and comparison with motif-based methods. a The AA composition of known degrons, Degpred degrons, and the human proteome, AAs are grouped based on their properties. b–d Distributions of predictions of intrinsic disorder, molecular recognition feature, and accessible surface area of predicted degrons in Degpred (blue), Motif_RF (orange), ELM motif matching (green), and random peptides from human proteome (gray, see Methods for detail). The intrinsic disorder was predicted with ESpritz-DisProt; the molecular recognition feature was predicted with MoRFchibi; the accessible surface area was predicted with ASAquick. P-values were calculated using two sides T-test. e–g Percentages of lysine, Ub-site, and Ub-lysine in the flanking 50 AAs of predicted degrons in Degpred (blue), Motif_RF (orange), and ELM motif matching (green), averages of the human proteome are displayed in gray. Both sides of predicted degrons were considered