Abstract
The recent outbreak of the novel corona virus disease 2019 (COVID-19) has been made a serious global impact due to its high infectivity and severe symptoms. The Severe Acute Respiratory Syndrome (SARS-CoV-2) RNA extraction is considered as one of the most important steps in COVID-19 detection. Several commercially available kits and techniques are currently being used for specific extraction of SARS-CoV-2 RNA. However, such methods are time consuming and expensive due to the requirement of trained labors, and several chemical reagents. To overcome the mentioned limitations, magnetic RNA adsorption methodology of glycine functionalized iron oxide nanoparticles (GNPs) was established. It showed an efficient potential in SARS-CoV-2 RNA extraction due to pH responsive nature of GNPs. The highly magnetic pH responsive GNPs were synthesized by one-pot co-precipitation method. Random morphology and average 20 nm size of GNPs were denoted by Transmission Electron Microscopy (TEM). X-ray diffractometer (XRD) showed the crystalline magnetite nature. Fourier transform infrared spectroscopy (FT-IR) and UV–visible spectrometry confirmed the presence of glycine on the surface of magnetic nanoparticles. Furthermore, the magnetic nature and thermal properties of GNPs were examined by vibrating sample magnetometer (VSM) and thermo-gravimetric analysis (TGA), respectively. In this study, glycine performed the role of RNA adsorbent. The adsorption of RNA onto the surface of GNPs was achieved in acidic medium (pH 6). In contrary, the elution of RNA from the surface of GNPs was achieved in basic medium (pH 8). The purity of obtained RNA was analyzed by UV–visible spectrometry. Further, the obtained RNA was examined for the presence of SARS-CoV-2 specific Envelope (E), RNA dependent RNA polymerase (RDRP) and Nucleocapsid (N) genes using an RT-PCR analysis. It showed the sudden rise in amount of these genes after 25 cycles of RT-PCR and hence indicated the efficient RNA extraction by GNPs. Agarose gel electrophoresis was used for validation of the quantity and quality of RNA extracted from SARS-CoV-2 patient’s sample. The reusability studies of GNPs were performed by monitoring the repeated use of GNPs for SARS-CoV-2 RNA extraction. This method possesses potential role in the field of disease diagnosis. The extraction results of RNA from SARS-CoV-2 patient’s sample indicated that the GNPs have an outstanding property over the current existing extraction protocols. It leads to the new advancements in extraction and detection of RNA.
Graphical Abstract
Graphical abstract of the pH responsive SARS-CoV-2 RNA extraction by using glycine functionalized magnetic iron oxide nanoparticles (GNPs) which were prepared by modified cost effective one pot chemical synthesis method. Prepared GNPs were characterized by XRD, FT-IR and UV-Visible spectrometry, Scanning electron microscopy (SEM) and Transmission electron microscopy (TEM). Glycine present on the surface of nanoparticles (NPs) played an important role in pH responsive RNA extraction procedure. When nanoparticles added in acidic (pH < 7) medium, glycine gained positive surface charge hence overall surface charge of NPs became positive. Thereby SARS-CoV-2 RNA adsorption/binding occurred on the surface of GNP. Later, the RNA-GNP complex was separated by an external magnet. Separated complex was added in basic (pH > 7) medium to elute RNA from GNP. This phenomenon occurred due to surface negative charge of glycine that caused charge repulsion with RNA. Eluted RNA was examined qualitatively and quantitatively by RT-PCR, nanodrop technique and agarose gel electrophoresis. Results were compared with kit based extracted RNA.

Supplementary Information
The online version contains supplementary material available at 10.1007/s10853-022-07464-6.
Introduction
The preliminary step of novel Corona Virus Disease 2019 (COVID-19) pandemic management is detection. Precise detection is achieved by the Reverse transcriptase-polymerase chain reaction (RT-PCR) technique. Currently, Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) RNA is extracted from the clinical sample by using commercial RNA extraction kit. The extracted RNA is processed further for the RT-PCR-based amplification of SARS-CoV-2 specific genes; nucleocapsid (N), envelope (E), and housekeeping in RT-PCR for confirming SARS-CoV-2 infection [1, 2].
Most of the commercially available RNA extraction kits rely on organic solvents which may pose challenge of false readout in PCR results. Moreover, the number of RNA extractions that can be carried out is also limited. The methodology also includes centrifugation cycles, and hence, a sophisticated lab set-up is necessary. Magnetic nanoparticle-based RNA extraction protocols offer a simple, economic and time saving approach of RNA extraction. Magnetic property of magnetic nanoparticles offers the tunable approach for RNA separation. The surface functionalization of magnetic nanoparticles reduces particle toxicity, increases particle surface area, provides surface charge and eventually makes particles highly efficient for RNA extraction [3–7]. The surface functionalization of magnetic nanoparticles with natural biopolymers (dextrin, silica, and albumin) offers a green sustainable approach. The bio-compatible biopolymers are non-toxic and presents array of advantages for downstream application. The biopolymer functionalized magnetic nanoparticles have been used as catalysts in different reactions. A new generation of magnetic nanoparticles with green functionalization of magnetic nanoparticles as an efficient, economic and environmental-friendly aspects to the experiments and hence widely been accepted by researchers [8–13].
The property of nucleic acid binding and elution by biopolymer functionalized magnetic nanoparticles has various biomedical functions such as gene transfection, biosensing, pathogen detection, and diagnosis of disease. Biopolymer functionalized magnetic nanoparticles are also used for biomolecule separation (DNA, RNA or protein) from real samples. Magnetic nanoparticle-based nucleic acid extraction methods are simple, economical, time saving, and ready for scale-up and can offer potential substitute to commercially available extraction kits [14–16]. In-spite of numerous advantages, there are limitations which needs to be addressed for validation of magnetic RNA extraction methodology at large scale. Herein, a simple and efficient pH-responsive SARS-CoV-2 RNA extraction methodology is presented. This ready for scale-up method offers the performance equivalent to commercial kits. Table 1 presents the comparison of our work with recently published work on nanocomposites for extraction of SARS-CoV-2 RNA.
Table 1.
Literature on extraction of SARS-CoV-2 RNA using magnetic nanocomposites and their limitations, sensitivity, reproducibility and specificity
| Sr no. |
Nanoparticle | RNA | Sample | Limitations | Sensitivity | Reproducibility | Specificity | Ref |
|---|---|---|---|---|---|---|---|---|
| 1 | Magnetic nanoparticles functionalized with poly amino ester with multiple carboxyl groups | SARS-CoV-2 | Pseudovirus | Chemicals like ammonium hydroxide (toxic), tetraethyl orthosilicate (TEOS), (3-Aminopropyl)triethoxysilane (APTES), and dimethyl sulfoxide (DMSO), 1,4-butanediol diacrylate, 6-amino caproic acid, sodium chloride (NaCl), sodium iodide (NaI), tri(hydroxymethyl)aminomethane (Tris), ethylene diamine tetraacetic acid (EDTA) and polyethylene glycol 8000 have been used. Hence overall cost exceeded | 2 μg of RNA was absorbed by 20 μg of synthesized MNPs. More than 90% RNA was absorbed | Not specified | The RNA extraction and detection were specific to the N gene of SARS-CoV-2 | [17] |
| 2 | Magnetic nanoparticles functionalized with poly amino ester with multiple carboxyl groups | SARS-CoV-2 | - | A systematic physicochemical characterization protocol had been carried out of nanoparticles which can be useful for SARS-CoV-2 RNA extraction. RT-PCR of extracted RNA had been performed. However, confirmatory results of different experiment sets are lacking in the study | 5, 10, 25, and 40 μL synthesized nanoparticles were mixed with the nasopharyngeal swab. Sensitivity was observed after the 38th cycle of RT-PCR | Not specified | The RNA extraction and detection were specific to N, E, and RDRP genes of SARS-CoV-2 | [18] |
| 3 | Magnetic nanocomposites functionalized with chitosan and graphene oxide | SARS-CoV-2 RNA | - | A protocol for magnet-based separation of SARS-CoV-2 RNA had been proposed. However, implementation of the above-mentioned protocol has been lacking | Not specified | Not specified | Not specified | [19] |
| 4 | Magnetic beads | SARS-CoV-2 RNA | Clinical sample | Fluorescence instrumentation setup, RT-LAMP primers, and instrumentation has been highly required. Moreover, the reusability of beads has not been mentioned or addressed | 100% sensitivity was obtained for E gene-specific SARS-CoV-2 virus | Not specified | The RNA extraction and detection were specific to the E gene of SARS-CoV-2 | [20] |
| 5 | Glycine functionalized iron oxide nanoparticles | SARS-CoV-2 | Clinical sample | Still need to be validated for commercial purpose | 100% sensitivity was obtained for E, N and RDRP gene-specific SARS-CoV-2 virus | The experiment supports the reusability of GNPs thrice for the extraction of SARS-CoV-2 | The RNA extraction and detection were specific to the E, N and RDRP gene of SARS-CoV-2 | Present study |
Glycine functionalized magnetic iron oxide nanoparticles (GNPs) were synthesized by the co-precipitation one-pot synthesis. No other binders like APTES were used for enhancing the binding interaction between nanoparticle and glycine. Glycine has pH-responsive nature, hence acquires positive surface charge in acidic (< 7) and negative surface charge in basic (> 7) media which allows adsorption and elution of SARS-CoV-2 RNA on and from GNPs according to the pH change, respectively. Magnetic property of GNPs provides the magnet-based rapid separation of RNA. Additionally, the same GNPs can be reused thrice for SARS-CoV-2 RNA extraction. Hence, an economical, eco-friendly, cost-effective and simple GNP-based SARS-CoV-2 RNA extraction methodology is established.
Materials and methods
Materials
Iron chloride hexahydrate (FeCl3.6H2O), iron chloride tetrahydrate (FeCl2.4H2O), sodium hydroxide (NaOH), sodium chloride (NaCl), ethylenediaminetetraacetic acid (EDTA), tris buffer, glycine, agarose, ethidium bromide (EtBr), triacetate EDTA buffer (TAE), DNA ladder, and loading dye were purchased from Himedia laboratories Pvt Ltd. Additionally, absolute ethanol, potassium bromide (KBr) were purchased from Sigma Aldrich Pvt ltd. RNA extraction kit and RT-PCR kit were purchased from MB615 Himedia Pvt Ltd, and MBPCR243 Himedia Pvt Ltd, respectively.
One-pot synthesis of GNPs
GNPs were synthesized using the co-precipitation one-pot synthesis. FeCl3.6H2O and FeCl2.4H2O were used as a precursor solution in 2:1 ratio and sodium hydroxide (NaOH) as a reducing agent. 0.016 M glycine solution was directly added to a previously prepared precursor solution. The mixture was vigorously stirred at 60 °C for 15 min. 2 M NaOH solution was added dropwise till the black precipitate appeared. pH was maintained continuously at 12. The suspension was kept at 60 °C for 6 h on a magnetic stirrer. The precipitate was separated by an external magnet. It was washed thrice with sterile distilled water and desiccated at 60 °C in an oven [21]. Obtained GNPs were crushed into powder and used for further operation.
Physiochemical characterization of GNPs
TEM examined the morphology and average particle size of synthesized GNPs. The crystalline nature of GNPs was determined by an XRD data. Results were documented with an XRD instrument (Rigaku miniflex diffractometer) using Cu K alpha radiation where K alpha = 1.542 A° and Bragg angle stretches from 5 to 80°. UV–visible spectrometry (Cary 60 UV–Vis spectrometer) and FT-IR confirmed the presence of glycine on the magnetic nanoparticles. UV–visible spectrometry displayed absorption spectra in the range of 200–600 nm. The FT-IR displayed the data in the 4000 to 400 cm−1. Further, TGA (SDT Q600 V20.9 Build 20) data provided thermal properties of GNPs by heating under furnace at the range of 0–800 °C. Furthermore, VSM investigated the magnetic nature of GNPs.
pH-responsive SARS-CoV-2 RNA extraction by GNPs
SARS-CoV-2 RNA samples were collected from the patient's nasal swabs by conventional RNA extraction kit (MB615–Hipor A viral RNA extraction kit–Himedia Pvt. Ltd.) in a viral transport medium and were stored at – 20 °C for further investigation. Standard safety procedures for COVID-19 were followed throughout the experiments. Personal protective equipment (PPE) kits were used during the study. No direct exposure to the SARS-CoV-2 virus was experienced. RT-PCR samples, extracted RNA, clinical samples, and laboratory waste were segregated in separate yellow-colored plastic bags with a symbol of cytotoxicity. Samples and used materials were discarded as per biomedical waste management policy.
pH-responsive GNP-based RNA extraction methodology was divided in five steps; binding of RNA on GNPs, magnetic separation of RNA-GNP complex, washing of complex, elution of RNA from GNPs and collection of the eluted RNA. Binding buffer comprised 25 mM Ethylenediaminetetraacetic acid (EDTA), 3 M sodium chloride (NaCl), and adjusted to pH 6. Washing buffer comprised 75% ethanol. Elution buffer comprised 0.5 M Tris buffer powder and adjusted to pH 8. All buffers were prepared in double distilled water.
Swab sample (200 µL) was incubated with the binding buffer containing GNPs (50 μg) for 10 min at room temperature on a shaker set to 500 rpm. The formed RNA-GNP complex was separated by an external magnet. The complex was washed with washing buffer thrice to remove excessive binding and impurities. Further, the RNA was separated from GNPs by adding an elution buffer at 55 °C under vigorous shaking for 10 min [17, 22]. The eluted RNA was collected separately and stored at–20 °C for further experiments.
Validation of RNA extraction
Preliminary analysis of GNP-based RNA extraction was carried out by UV–visible spectrometry (Nanodrop ThermoScientific multiskan sky). The eluted RNA was examined for A260/A280 RNA purity. Further, RT-PCR analysis was performed for the qualitative and quantitative estimation of extracted RNA. SARS-CoV-2 specific primers were added to the reaction mixture according to the manufacturer's instructions. RT-PCR was performed with a HiPCR coronavirus (COVID-19) multiplex probe PCR kit (MBPCR243 Himedia Pvt. Ltd). 40 thermal cycles were carried out where cDNA synthesized for 15 min at 50 °C, followed by initial denaturation for 3 min at 95 °C, 40 cycles of 15 s at 95 °C, and 30 s at 58 °C. The obtained RT-PCR products (cDNA) were confirmed qualitatively by an agarose gel electrophoresis. RNA extracted by conventional RNA extraction kit was considered a positive control.
Reusability of GNPs
In order to check the reusability of GNPs, RNA adsorption and elution process was repeated 3 times by providing same set of GNPs. Every experiment was done in triplicates to validate its reproducibility.
Statistical analysis
All experiments were carried out six times (N = 6). Statistical data were obtained with standard deviations.
Results and discussion
GNP synthesis and characterization
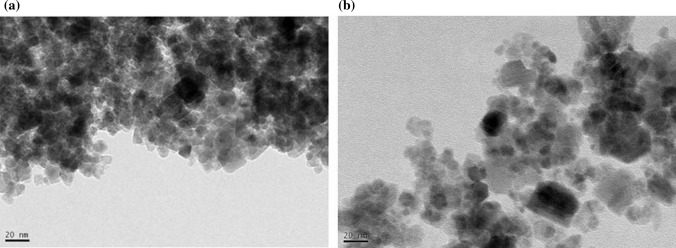
GNPs were synthesized by co-precipitation one-pot synthesis in presence of amino acid glycine as a biomolecule shown in Fig. 1. Glycine molecule possesses one hydroxyl group (OH) and iron oxide nanoparticle possesses two hydrogen atoms. Hydroxyl groups of two glycine molecules form bonds with hydrogen atoms of Fe3O4. NH2 group of glycine remains available for further RNA interactions. TEM images showed the morphology of particles in Fig. 2. The average size of particle was observed as 20 nm. Morphologically GNPs showed similarity with bare iron oxide nanoparticles. However, aggregation of particles was reduced in GNPs when compared with the bare iron oxide nanoparticles. The physicochemical characterization of synthesized GNPs is shown in Fig. 3. The XRD pattern of bare iron oxide nanoparticles (IONPs) and GNPs is shown in Fig. 3a. The diffraction patterns were well matched with magnetite (JCPDS card number 75–0449) confirming the formation of Fe3O4. It confirms the crystalline nature of GNPs. The appearance of peak 311 indicates the presence of magnetite phase of iron oxide in GNPs. The absence of 210 and 300 in both patterns rules out the possibility of the presence of separate maghemite (gFe2O3) in the sample [13]. Interestingly, the XRD patterns of GNP showed additional sharp diffraction peaks (marked by *) at 300 and 302 which was not present in bare IONPs. These relatively sharper diffraction peaks may be attributed by the presence of glycine in GNPs [23]. It is noteworthy that the addition of glycine did not affect the diffraction pattern of bare IONPs suggesting the successful surface functionalization of glycine onto the Fe3O4.
Figure 1.
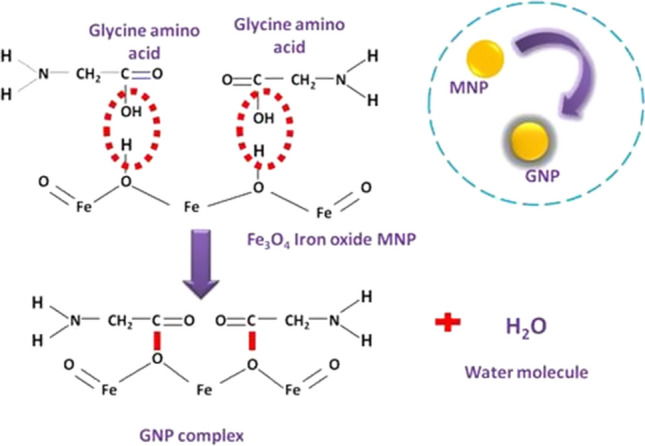
Proposed chemical binding of glycine on the surface of magnetic iron oxide nanoparticles. Glycine molecule possesses one hydroxyl group (OH) and iron oxide nanoparticle possesses two hydrogen atoms. Hydroxyl groups of two glycine molecules form bonds with hydrogen atoms of Fe3O4
Figure 2.
Morphology of synthesized GNPs; a TEM of bare iron oxide nanoparticles, b TEM of GNPs suggesting average 20 nm size
Figure 3.
Physicochemical characterizations of synthesized GNPs; a XRD suggesting crystalline magnetite nature, b ATR-IR suggesting presence of glycine, c TGA suggesting thermal properties, d VSM analyzing magnetic nature
FTIR study was undertaken to confirm the chemical bonds between magnetic Fe3O4 NPs and glycine. Figure 3b displays the FTIR spectrum of GNP within the wavenumber 4000–400 cm−1. The presence of peak at 580 to 680 cm−1 indicated the metal-oxygen bond occurred due to Fe–O [24]. The bands at 846 and 1061 cm−1 are due to amine groups present in the glycine. NH bending is indicated by the band at 1335 cm−1. OH and NH overlapping vibrations are responsible for the bands at 1622 and 3300 cm−1, respectively [25]. In addition to this, the thermal properties of the GNPs were also examined using TGA technique in the temperature range of 35–800 °C in a nitrogen atmosphere. As shown in Fig. 3c, the GNPs are stable below temperature 200 °C with no significant weight loss. The initial weight loss (around 7%) occurred due to the evaporation of water content present in the NPs [26]. A continues weight loss was observed within the temperature 200–600 °C where sudden weight loss within the temperature range 210–270 °C was due to sublimation and decomposition of surface coated glycine [27]. Upto 600 °C, almost 56% weight loss took place. It is well known that, Fe3O4 NPs are highly stable over wide range of temperature, it can be said that the weight loss observed in this case is only because of the decomposition of glycine. VSM was performed to study magnetic properties of GNPs as shown in Fig. 3d. Hysteresis loop demonstrated superparamagnetic properties of GNPs. The magnetization saturation of GNPs was observed at 45 emu·g − 1. Strong magnetization property of GNPs is critical for the magnet-based biomolecule extraction.
Functionalization of GNPs was further examined by UV–Visible spectrometry as shown in Figure S1. Maximum absorbance was obtained at 210 nm showing the presence of amino acid glycine. A small absorption at 360 nm indicated the presence of Fe3O4 NPs.
pH-responsive SARS-CoV-2 RNA extraction by GNPs
pH-responsive SARS-CoV-2 RNA extraction by using GNPs was performed by a simple and cost-effective protocol. Basically, in this catalytic study where amino acid–glycine acted as a mediator for efficient SARS-CoV-2 RNA extraction. From the obtained characterization data, it was observed that the GNPs possess nearly spherical morphology and it was interpreted that the glycine organic chains were covering the surface of magnetic nanoparticles in resemblance with the fishing net that captures the prey from its vicinity. Glycine amino acid exists as zwitterion at neutral pH, cation in acidic pH, and anion in basic pH described in Fig. 4a. This powerful pH influence of the charge state was reflected to change their properties and sensitivity towards the biomolecules (specifically RNA) according to the provided environment. Such changing pH conditions alter their charge and extract and elute charge-specific biomolecules, respectively, by simple attractive and repulsive forces, which in turn gained dramatic effect on specific biomedical activity. Carbon (C) and Oxygen (O) present in the organic chain of glycine were responsible for the charge state. In this study, three buffer solutions were prepared to provide different pH-responsive conditions to glycine as demonstrated in Fig. 4b. Buffer 1 was named as a binding buffer and gained the acidic pH. On the contrary, buffer 2 named elution buffer gained the basic pH. The third buffer was named washing buffer and it was pH-independent. The process of SARS-CoV-2 RNA extraction was divided into two sub-processes from which the first was the formation of the RNA-GNP complex and the second was the elution of RNA from the GNPs. The process of RNA-GNP complex formation was performed in binding buffer media. At acidic pH, GNPs became maximally protonated and gained a positive surface charge due to the presence of NH3+. This protonated form became responsible for attracting RNA present in the clinical sample towards the GNP as RNA possesses a strong de-protonated phosphate backbone in its structure. The overall charge of RNA is negative due to the presence of a phosphate backbone. Hence, negatively charged RNA was attracted towards the positively charged GNPs. As the viral body possesses specific RNA and the envelope proteins (which are mostly possessing a neutral surface charge), it became easier to separate the RNA by changing the charge state. Thereby, the RNA-GNP complex was formed in the binding buffer. Later, the complex was separated from the buffer by external magnet as the GNPs were strongly magnetic. The important step was washing where the complex was given a wash thrice with the washing buffer to remove the charge-independent biomolecules (other than RNA) from the complex. It was observed that, as the washing buffer was neutral in pH it could not separate RNA from GNP but the other non-specific biomolecules. Moving forward to the process of RNA elution, the complex was mixed with the elution buffer which had basic pH (< 7). Hence, glycine present on the GNPs became maximally de-protonated and gained a negative surface charge due to the presence of CO2−. This de-protonated form became responsible for repelling or eluting RNA present in the RNA-GNP complex. Hence, negatively charged RNA was eluted from the negatively charged GNPs due to charge repulsion. The GNPs were separated from RNA by an external magnet and reused for further RNA extraction from other clinical samples.
Figure 4.

pH-responsive SARS-CoV-2 RNA extraction by GNPs; a pH-dependent RNA binding and elution, b GNP-based methodology
Validation of RNA extraction
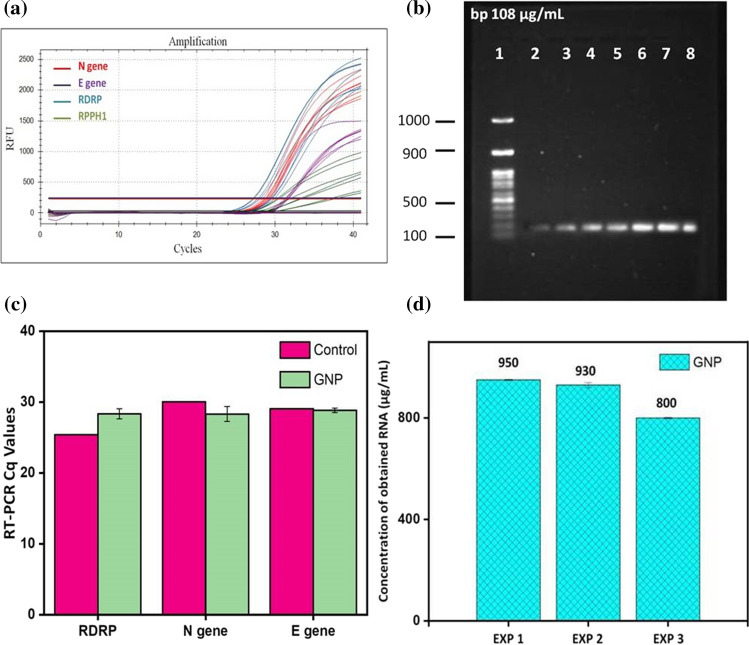
The significant extraction efficiency of GNPs under pH influence was observed when compared with a conventional kit-based SARS-CoV-2 RNA extraction. By establishing an easy protocol of binding and elution of RNA on and from GNPs, respectively, as described in Fig. 4b, RNA was extracted from a clinical sample. The purity of viral RNA (A260/A280) extracted by GNPs was equivalent to kit-based extraction as described in Figure S2 and Table S1 (supplementary). It was observed that the RNA purity obtained by GNP mediation was similar to that of a conventional commercial kit. Hence, a study was further processed for confirmatory tests. Extracted RNAs were amplified by performing RT-PCR analysis and results were compared with RNA extracted by a conventional commercial kit. Orderly equivalent data were obtained for GNP mediated extraction when compared with RNA extracted by kit, as indicated in Fig. 5a and c. RT-PCR was carried out for six GNP extracted RNA samples and compared with positive (kit extracted RNA) as well as negative control (sample without RNA). Nucleocapsid (N) gene, Envelope (E) gene, and RNA dependent RNA polymerase (RDRP) gene of SARS-CoV-2 were targeted to obtain SARS-CoV-2 RNA amplification and to check the efficiency of extraction from the sample. The sudden rise in peaks for each gene after 25 RT-PCR cycles indicated the presence and amplification of SARS-CoV-2 RNA. Quantification of the GNP-mediated RNA extraction is shown in Table S2 (supplementary).
Figure 5.
pH responsive RNA extraction using magnetic GNPs; a RT-PCR gene amplification curve suggesting the presence of SARS-CoV-2 genes in extracted RNA, b Agarose gel electrophoresis of obtained RT-PCR product, c quantitative estimation of SARS-CoV-2 specific genes amplified by RT-PCR, d reusability study of pH-responsive GNPs. Data = Mean ± SD. N = 6
For further confirmation, the cDNA of SARS-CoV-2 was analyzed by 1.2% agarose gel electrophoresis previously stained with EtBr. Significant cDNA bands could observe on an agarose gel as illustrated in Fig. 5b. The overall surface charge of nucleic acid is negative due to its phosphate backbone. cDNA extracted by GNPs from 6 samples were loaded into the well towards the end of a cathode (negatively charged electrode) and provided voltage (50 V) and current (30A). Once the voltage has been provided, cDNA with a negative charge runs towards the anode (positively charged electrode) on an agarose gel matrix. Ethidium bromide (EtBr), an intercalating dye was added to the agarose gel before its solidification. It intercalated a nucleic acid (cDNA) and fluoresced. Hence, fluorescence was observed at the respective location of cDNA on a gel. The whole electrophoresis assembly was submerged in 1X TAE buffer to maintain pH conditions.
Reusability of the GNPs
The efficiency to reuse and recycle magnetic nanomaterials for biomedical purposes is one of their beneficial aspects when considered economical and environmental views. Studies on reusability of GNPs were carried out by analyzing the concentration of SARS-CoV-2 RNA obtained from COVID-19 positive clinical sample by using same set of GNP nanoparticles for three subsequent experiments as shown in Fig. 5d. 950 µg RNA was obtained from the first experiment from 1 mL of sample. The same set of GNPs incorporated for the second experiment and, 930 µg of RNA concentration was obtained. The obtained concentration in the second experiment was only 2% lower than the first one. Likewise, the third experiment provided 800 µg RNA from the same set of GNPs. It refers that even after repeated incorporation of GNPs, the binding and eluting properties were not reducing significantly. It indicates the SARS-CoV-2 RNA extraction ability of GNPs with changing pH is remarkable and they remain highly stable; hence become suitable candidate for repeated RNA extraction.
Conclusions
In this study, a cost-effective simple, and efficient pH-responsive SARS-CoV-2 RNA extraction method by using amino acid glycine functionalized magnetic iron oxide nanoparticles (GNPs) was established. GNPs were developed by a one-pot chemical synthesis method. Glycine present on the surface of nanoparticles played an important role in the whole process and acted as a mediator for RNA binding. It provided a larger surface area and charge for the efficient binding of RNA on the particle surface. As the surface charge of GNPs depended on the pH of the reaction solution, binding and elution of RNA on and from the surface of GNP became achievable, respectively. In acidic pH, amino acid glycine gained a positive charge, and hence negatively charged RNA binding occurred. However, elution of RNA from GNP occurred in basic pH due to repulsive charges. Binding and elution buffers were prepared with minimum chemicals. Thus, whole extraction process became cost-efficient and simple. RNAs extracted from this method were efficiently detected by RT-PCR. Moreover, obtained RT-PCR products (cDNA) could efficiently show bands by agarose gel electrophoresis. The results were compared with the RNA extracted by commercial RNA extraction kit. Equivalent results were obtained for GNP extracted RNA. Hence, the pH-responsive GNP-based SARS-CoV-2 RNA extraction process could be potent for the detection of COVID-19. The kit is still to be validated for commercial use.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors have appreciated the support from the Intramural University project; “Targeted destruction of cancer stem cells by surface-functionalized magnetic nanoparticles” (project no. DYPES DUR&D/2021/274), D. Y. Patil Education Society, Kolhapur, India.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kevadiya BD, Machhi J, Herskovitz J, Oleynikov MD, Blomberg WR, Bajwa N, Gendelman HE. Diagnostics for SARS-CoV-2 infections. Nat Mater. 2021;20:593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J, Liu J, Li S, Peng Z, Xiao Z, Wang X, Luo J. Detection and analysis of nucleic acid in various biological samples of COVID-19 patients. Travel Med Infect Dis. 2020;37:101673. doi: 10.1016/j.tmaid.2020.101673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu H, Fu L, Jin Y, Shao J, Zhang S, Zheng N, Ye H. Clinical features of COVID-19 convalescent patients with re-positive nucleic acid detection. J Clin Lab Anal. 2020;34:23392. doi: 10.1002/jcla.23392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esbin MN, Whitney ON, Chong S, Maurer A, Darzacq X, Tjian R. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020;26:771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiwari AP, Rohiwal SS (2019) Synthesis and bioconjugation of hybrid nanostructures for biomedical applications. In: Hybrid Nanostructures for Cancer Theranostics pp 17–41
- 6.Chaimayo C, Kaewnaphan B, Tanlieng N, Athipanyasilp N, Sirijatuphat R, Chayakulkeeree M, Horthongkham N. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virol J. 2020;17:1–7. doi: 10.1186/s12985-020-01452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olivier V, Delphine M, Olivier R. Considerations for diagnostic COVID-19 tests. Nat Rev Microbiol. 2020;19(3):171–83. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eivazzadeh KR, Radinekiyan F, Maleki A, Salimi BM, Azizi M. A new generation of star polymer: magnetic aromatic polyamides with unique microscopic flower morphology and in vitro hyperthermia of cancer therapy. J Mater Sci. 2020;55:319–336. doi: 10.1007/s10853-019-04005-6. [DOI] [Google Scholar]
- 9.Maleki A, Hassanzadeh AF, Varzi Z, Esmaeili MS. Magnetic dextrin nano biomaterials: an organic-inorganic hybrid catalyst for the synthesis of biologically active polyhydroquinoline derivatives by asymmetric Hantzsch reaction. Mater Sci Eng C. 2020;109:110502. doi: 10.1016/j.msec.2019.110502. [DOI] [PubMed] [Google Scholar]
- 10.Maleki A. Green oxidation protocol: Selective conversions of alcohols and alkenes to aldehydes, ketones, and epoxides by using a new multiwall carbon nanotube-based hybrid nanocatalyst via ultrasound irradiation. Ultra sonochem. 2018;40:460–464. doi: 10.1016/j.ultsonch.2017.07.020. [DOI] [PubMed] [Google Scholar]
- 11.Maleki A. An efficient magnetic heterogeneous nanocatalyst for the synthesis of pyrazinoporphyrazine macrocycles. Polycycl Aromat Compd. 2018;38:402–409. doi: 10.1080/10406638.2016.1221836. [DOI] [Google Scholar]
- 12.Maleki A. Fe3O4/SiO2 nanoparticles: an efficient and magnetically recoverable nanocatalyst for the one-pot multicomponent synthesis of diazepines. Tetrahedron. 2012;68:7827–7833. doi: 10.1016/j.tet.2012.07.034. [DOI] [Google Scholar]
- 13.Tiwari AP, Rohiwal SS, Suryavanshi MV, Ghosh SJ, Pawar SH. Detection of the genomic DNA of pathogenic α-proteobacterium Ochrobactrum anthropi via magnetic DNA enrichment using pH responsive BSA@ Fe3O4 nanoparticles prior to in-situ PCR and electrophoretic separation. Microchim Acta. 2016;183:675–681. doi: 10.1007/s00604-015-1710-6. [DOI] [Google Scholar]
- 14.Xiao AT, Tong YX, Zhang S (2020) False‐negative of RT‐PCR and prolonged nucleic acid conversion in COVID‐19: rather than recurrence. J Med Virol [DOI] [PMC free article] [PubMed]
- 15.Tavallaie R, McCarroll J, Le GM, Ariotti N, Schuhmann W, Bakker E, Gooding JJ. Nucleic acid hybridization on an electrically reconfigurable network of gold-coated magnetic nanoparticles enables microRNA detection in blood. Nat Nanotechnol. 2018;13:1066–1071. doi: 10.1038/s41565-018-0232-x. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Wu Y, Chen Z, Hu Z, Fang Y, Liao P, He N. Performance evaluation of a novel sample in–answer out (SIAO) system based on magnetic nanoparticles. J Biomed Nanotech. 2017;13:1619–1630. doi: 10.1166/jbn.2017.2478. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Z, Cui H, Song W, Ru X, Zhou W, Yu X. A simple magnetic nanoparticles-based viral RNA extraction method for efficient detection of SARS-CoV-2. BioRxiv. 2020;382(12):727. [Google Scholar]
- 18.Chacón-Torres JC, Reinoso C, Navas-León DG, Briceño S, González G. Optimized and scalable synthesis of magnetic nanoparticles for RNA extraction in response to developing countries' needs in the detection and control of SARS-CoV-2. Sci Rep. 2020;10:1–10. doi: 10.1038/s41598-020-75798-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh V, Batoo KM, Singh M. Fabrication of chitosan-coated mixed spinel ferrite integrated with graphene oxide (GO) for magnetic extraction of viral RNA for potential detection of SARS-CoV-2. Appl Phys A. 2021;127:1–14. doi: 10.1007/s00339-020-04132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein S, Müller TG, Khalid D, Sonntag-Buck V, Heuser AM, Glass B, Chlanda P. SARS-CoV-2 RNA extraction using magnetic beads for rapid large-scale testing by RT-qPCR and RT-LAMP. Viruses. 2020;12:863. doi: 10.3390/v12080863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nosrati H, Salehiabar M, Attari E, Davaran S, Danafar H, Manjili HK. Green and one-pot surface coating of iron oxide magnetic nanoparticles with natural amino acids and biocompatibility investigation. Appl Organomet Chem. 2018;32:4069. doi: 10.1002/aoc.4069. [DOI] [Google Scholar]
- 22.Tiwari AP, Satvekar RK, Rohiwal SS, Karande VA, Raut AV, Patil PG, Pawar SH. Magneto-separation of genomic deoxyribose nucleic acid using pH responsive Fe 3 O 4@ silica@ chitosan nanoparticles in biological samples. Rsc Adv. 2015;5:8463–8470. doi: 10.1039/C4RA15806G. [DOI] [Google Scholar]
- 23.Azhagan SA, Ganesan S. Growth and characterization of gamma glycine single crystal from ammonium sulfate as solvent. Recent Res Sci Technol. 2010;2:6. [Google Scholar]
- 24.Lopez JA, González F, Bonilla FA, Zambrano G, Gómez ME. Synthesis and characterization of Fe3O4 magnetic nanofluid. Rev Latinoam de Metal y Mater. 2010;30:60–66. [Google Scholar]
- 25.Sheela NR, Muthu S, Krishnan SS. FTIR, FT Raman and UV-visible spectroscopic analysis on metformin hydrochloride. Asian J Chem. 2010;22:5049. [Google Scholar]
- 26.Awwad AM, Salem NM. A green and facile approach for synthesis of magnetite nanoparticles. J Nanosci Nanotechnol. 2012;2:208–213. doi: 10.5923/j.nn.20120206.09. [DOI] [Google Scholar]
- 27.Azhagan SA, Kathiravan VS. Selective crystallization of gamma glycine for NLO applications using magnesium sulfate (MgSO4) as an additive. Mater Sci Pol. 2019;37:2. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.