Figure 3. Identification of E1E2 amino acids critical for infectivity.
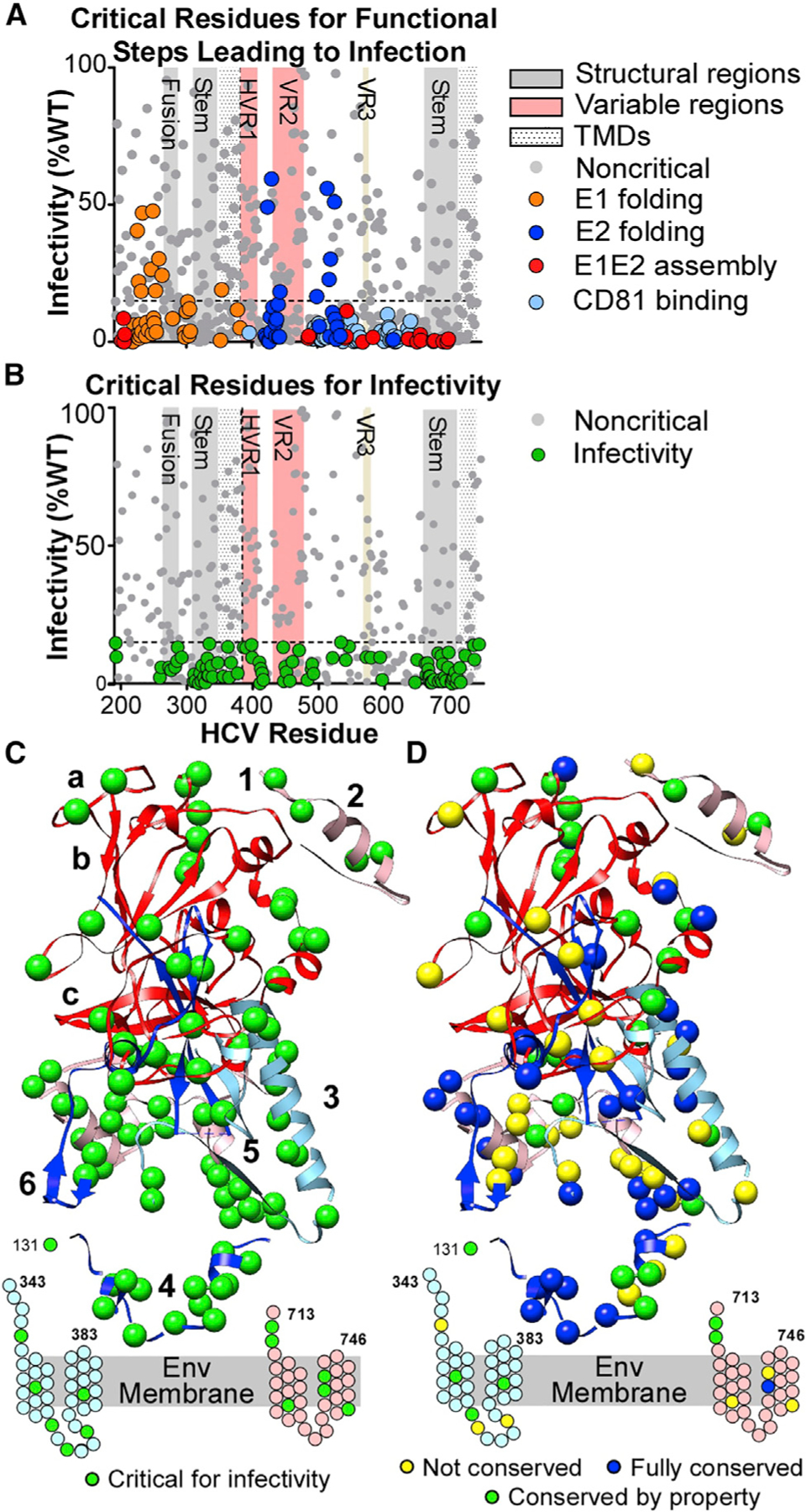
(A) Infectivity values (mean of 5 experiments) for E1E2 clones with mutations critical for E1 or E2 folding, E1E2 heterodimer assembly, and CD81 binding.
(B) Infectivity values (mean of 5 experiments) for clones not critical for functional steps (i.e., excluding clones colored in A). Green circles are critical residues for infectivity (clones with mean infectivity ≤15% and with ≥70% for E1E2 folding, E1E2 heterodimer assembly, and CD81-LEL binding). Gray circles below the threshold line represent clones with mean reactivity <70% of WT in previous functional steps. Important structural regions are marked, including HVR1, VR2, and VR3.
(C) Critical residues for infectivity are shown as green spheres on the E1E2 composite structure. Residues in the stem and TM regions of E1 and E2 that lack structures are shown as cartoons. Annotations are as in Figure 1A.
(D) Structural representation of E1E2 critical residues for infectivity, characterized by their conservation across the 7 HCV genotypes. Residues fully conserved across all 7 genotypes are shown as dark blue spheres. Residues with conserved properties are shown as green spheres. Non-conserved residues are shown as yellow spheres.
