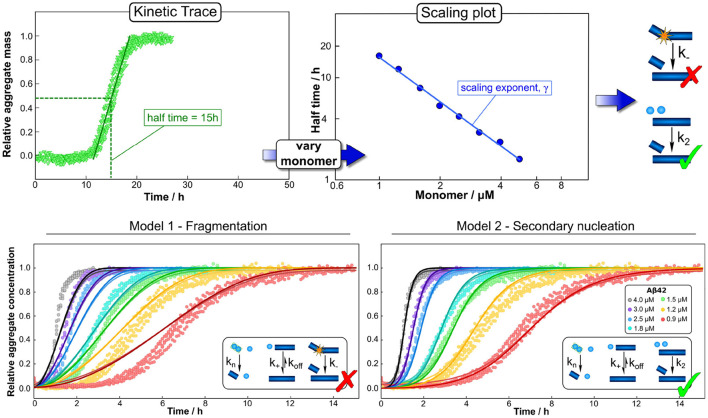
Figure 3.
Global fitting and the use of scaling to determine mechanisms. Varying a number of different experimental parameters is crucial for determining possible mechanisms. In particular, variations of the monomeric protein concentration can provide important mechanistic insights due to the differing dependence of different mechanisms on monomer concentration. A powerful and easy way to apply this technique in practice is the use of scaling exponents, which describe how a representative quantity of the aggregation reaction, such as the half time, varies with the monomer concentration. (Top) The half time can easily be extracted from kinetic traces. Plotting the half time against monomer concentration then allows the determination of the scaling exponent and exclusion of mechanisms that are inconsistent with the observed scaling. Often scaling exponents alone already allow the qualitative determination of mechanisms (here an example in which fragmentation can be excluded simply based on the scaling exponent). (Bottom) To then quantify the rates and confirm mechanisms, one performs a global fit, i.e., using one set of parameters to describe the entire dataset at all monomer concentrations, of the integrated rate laws derived for different models (points are experimental measurements of aggregate amounts, solid lines the best global fit of the model shown in the schematic, different colors denote different protein concentrations). Here the fits confirm the conclusions from the scaling analysis, that a fragmentation dominated mechanism is inconsistent with the data while a secondary nucleation dominated mechanism describe it well. Adapted from Meisl et al. (2016a, 2017b).