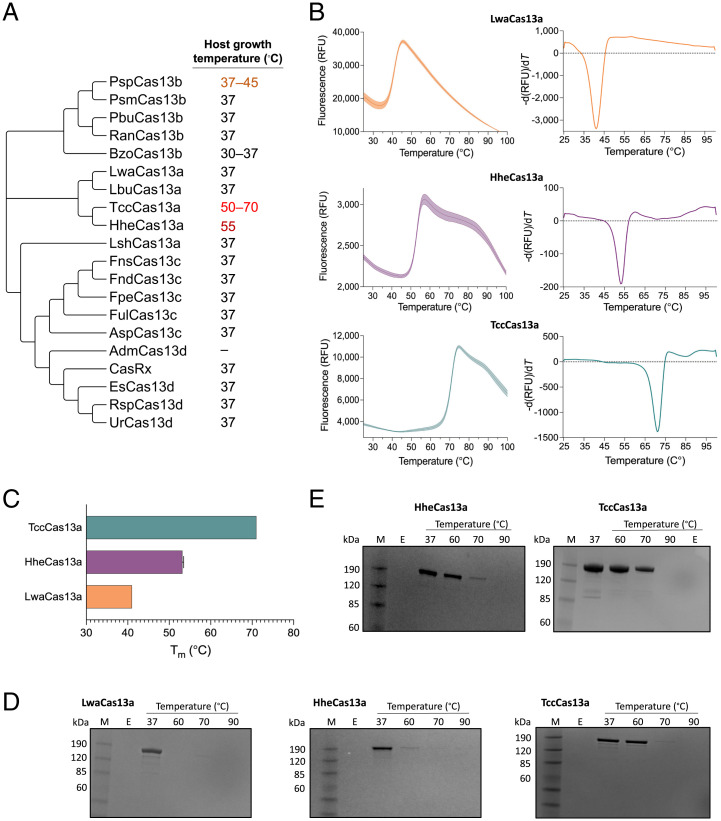
Fig. 1.
Thermostability analysis of thermophilic Cas13 effectors. (A) Maximum-likelihood phylogenetic tree of Cas13 proteins from different organisms. The tree was generated using MEGA X software. Most selected proteins were isolated from mesophilic bacteria, although several have been cultivated as thermophiles and thus offer an interesting collection of high temperature–stable proteins. TccCas13a and HheCas13a were selected as potentially thermophilic Cas13 proteins. (B) Differential scanning fluorimetry (DSF) profiles of protein melting points using a conventional real-time PCR instrument. The peak in the left graph indicates protein denaturation. The right-side graph is the derivative of the left-side graph. (C) Denaturation temperatures of TccCas13a, HheCas13a, and LwaCas13a proteins, as determined by DSF in B. Data are shown as mean ± SD (SD) (n = 3). (D) SDS-PAGE illustrating the protein stability of TccCas13a, HheCas13a, and LwaCas13a after incubation for 30 min at different temperatures. M, protein marker; E, empty well. (E) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) showing the protein stability of TccCas13a and HheCas13a RNPs assembled with their cognate crRNAs. After assembly with crRNAs, TccCas13a and HheCas13a RNPs were incubated for 30 min at different temperatures before electrophoresis. RFU, relative fluorescence unit.