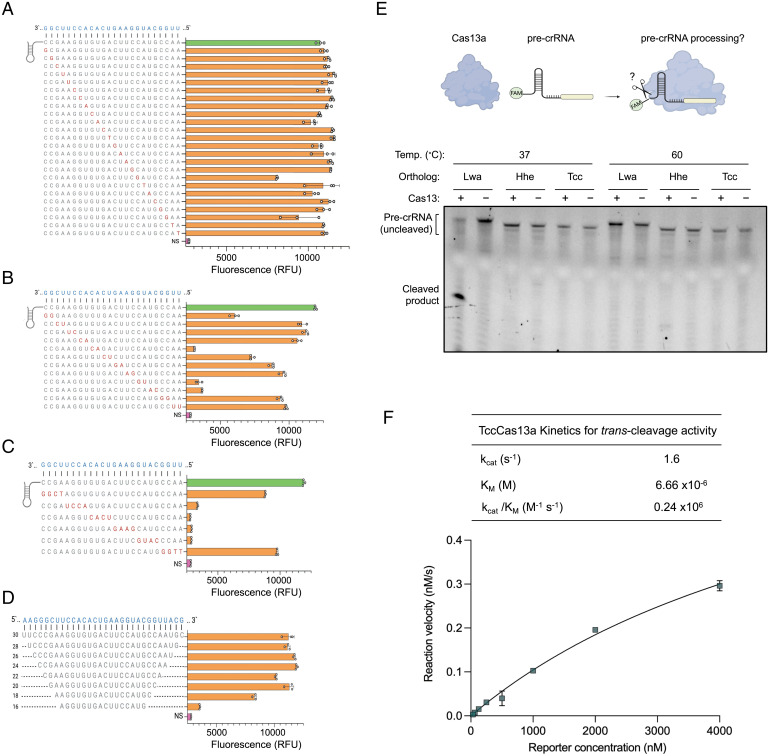
Fig. 3.
In vitro characterization of thermostable Cas13a crRNA sequence requirements. Evaluating the effect of single (A), double (B), and stretches of four mismatches (C) between crRNA and target RNA on TccCas13a activity. (Left) crRNA nucleotide sequence with the positions of mismatches (red) on the crRNA spacer. (Right) The fluorescence intensity, relative to the nonspecific crRNA control (NS) (pink) or crRNA with no mismatches (green), resulting from TccCas13a collateral cleavage activity on each tested crRNA. Reactions were incubated at 56 °C, and the end-point fluorescence signal was measured after 30 min. (D) TccCas13a RNA detection activity with different crRNA spacer lengths. (Left) crRNA spacer nucleotide sequences with the length of each spacer shown on the left of the sequence. (Right) The fluorescence intensity, relative to the NS (pink), resulting from TccCas13a collateral cleavage activity on each tested crRNA. Reactions were incubated at 56 °C, and the end-point fluorescence signal was measured after 30 min. (E) Representative denaturing gels showing Cas13a-mediated processing of their cognate precrRNAs; 100 nM each Cas13a ortholog was incubated with 200 nM cognate 5′-FAM–labeled precrRNA for 1 h at different temperatures. See SI Appendix, Fig. S5A for uncropped gel picture. (F) Representative Michaelis-Menten plot for TccCas13a-catalyzed ssRNA trans cleavage activity. Enzyme kinetic data and the measured Kcat, Km, and Kcat/Km are shown on the top of the plot. Values are shown as mean ± SD (n = 3). RFU, relative fluorescence unit.