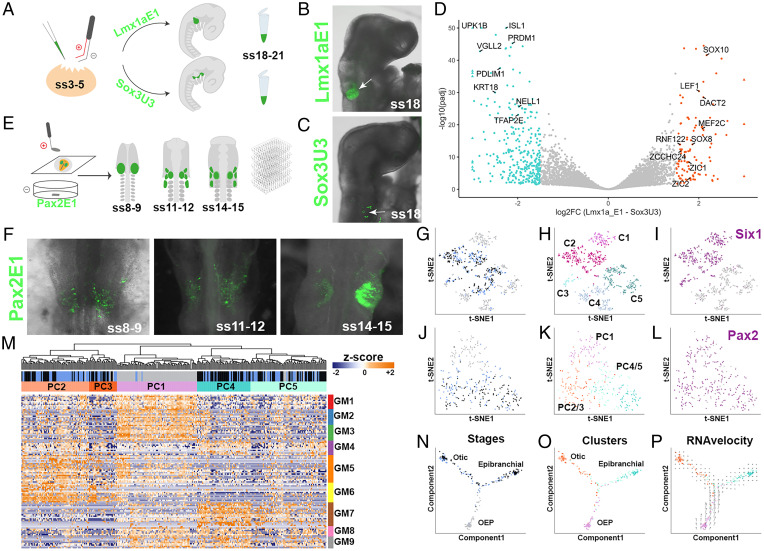
Fig. 1.
Transcriptomic characterization of ear development. (A–C) In ovo electroporation (A) was used to label and collect otic (B; Lmx1aE1-EGFP+) and epibranchial (C; Sox3U3-EGFP+) cells for bulk RNAseq. (D) Volcano plot showing genes differentially expressed (absolute log2 fold change > 1.5 and adjusted P value < 0.05) in otic (orange) and epibranchial cells (green). (E, F) Cells expressing the Pax2E1-EGFP reporter active in OEPs and otic and epibranchial placodes were collected for scRNAseq at the stages indicated (F). (G–L) t-distributed stochastic neighbor embedding (tSNE) representation of unsupervised hierarchical clustering of all cells (G–I) and of the placodal cell subset (J–L); cells are color-coded according to the stage collected (G, J; OEP gray, ss11-12 blue, ss14-15 black), clusters (H, K), and placodal marker expression: Six1 (I) and Pax2 (L). (M) Heatmap showing partitioning of the placodal subset (C1, C2 in H) into five clusters (PC1–5) based on gene modules (GM). Expression profiles reveal that PC1 largely contains OEP-like cells, while PC2/3 and PC4/5 are composed of otic-like and epibranchial-like cells. (N–O) Pseudotime ordering using Monocle2 shows trajectories between stages (N) and clusters (O). Note that otic- and epibranchial-like cells segregate into two branches: otic-like in orange, epibranchial-like in green, and OEP-like in pink. (P) RNA velocity vector field verifies the directional trajectories predicted by Monocle2.