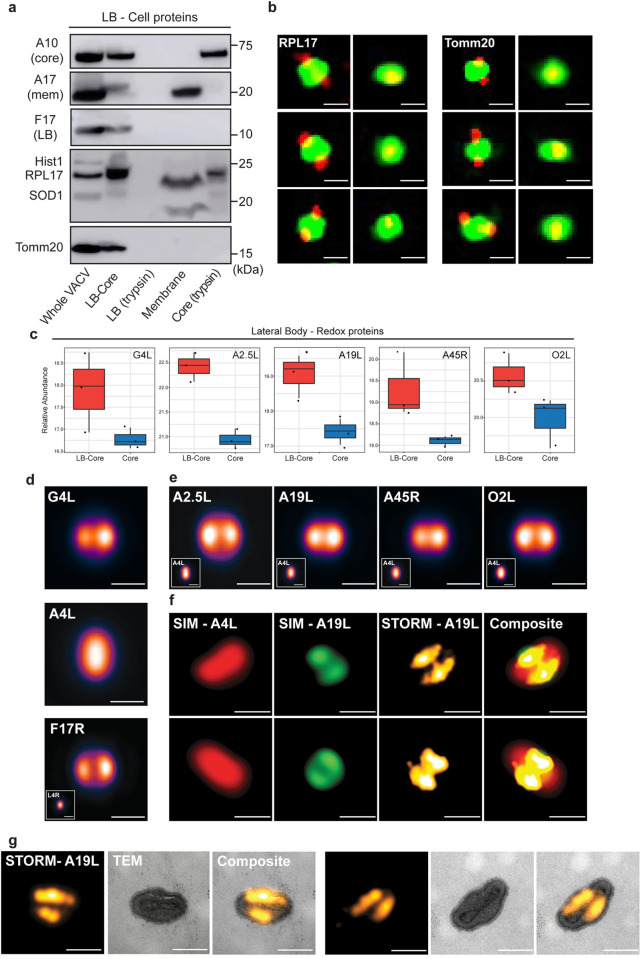
Fig 3. Cell and viral redox proteins reside in LBs.
(a) Biochemical localization of candidate cellular LB proteins. MVs were subjected to controlled degradation as outlined in Fig 1A, and fractions subjected to immunoblot analysis for indicated viral and cellular proteins. Experiment was performed in triplicate and representative blots displayed. (b) SIM images (sagittal and frontal orientation) of RPL17 and Tomm20 localization in fractionated WR EGFP-A4L virions. Experiments were performed in duplicate and six representative virions displayed. (c) Relative abundance plots of the five putative redox modulating viral enzymes (G4L, A2.5L, A19L, A45R, O2L) across LB-Core and Core fractions (d) VirusMapper models (sagittal orientation) of G4L (LB) and A4L (core) protein localization using WR mCherry-A4L G4L-EGFP virions (n = 949). A VirusMapper model of WR L4R-mCherry EGFP-F17R is displayed for comparison (n = 231) (e) VirusMapper models (sagittal orientation) of EGFP-tagged redox proteins: A2.5L, A19L, A45R, O2L (n > 100). (f) Correlative SIM-STORM imaging of WR mCherry-A4L A19L-EGFP virions. Two representative virions are displayed (Overview in S4A Fig) (g) Correlative STORM-TEM of WR mCherry-A4L A19L-EGFP virions immunolabelled with anti-GFP nanobody directly conjugated to AlexaFluor647-NHS. STORM images of A19L were registered with EM micrographs. Two representative virions are displayed (Overview S4B Fig). (b, d-f) Scale bars = 200 nm.