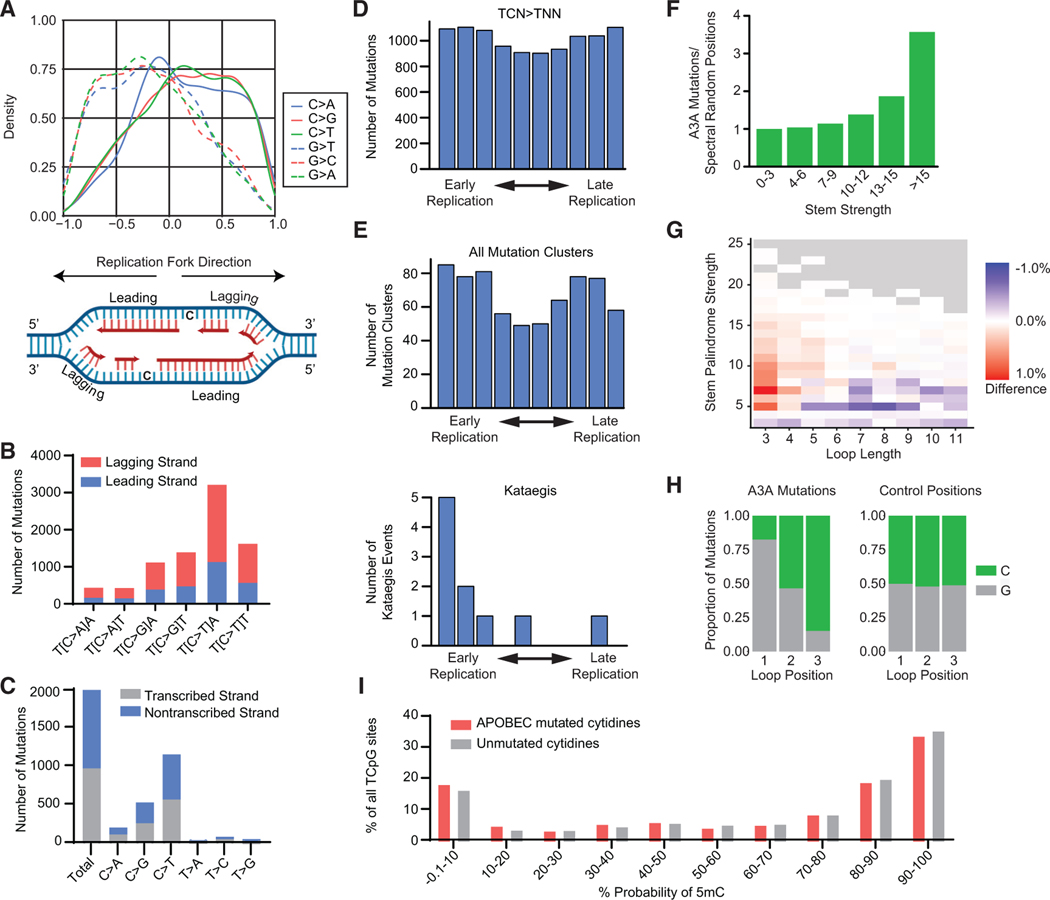
Figure 5. APOBEC3A deamination occurs at specific genomic substrates.
(A) Density plot of mutated C and G bases relative to replication fork directionality (RFD). Positive RFD indicates a rightward-oriented fork, in which case the sense strand is the lagging strand. Negative RFD indicates a leftward-oriented fork. RFD is calculated by the difference between rightward- and leftward-oriented forks; thus, a value of 0 means that, at that location, equal numbers of forks go to the right and to the left. A replication fork diagram depicting leading and lagging strand directions is shown below.
(B) SBSs found to be associated with replication forks from all DT40-A3A descendant clone genomes are quantified as leading-strand (blue) or lagging-strand(red) mutations. Base substitution in the indicated trinucleotide context is shown on the x axis.
(C) The numbers of SBSs in transcribed and non-transcribed strands from DT40-A3A genomes are quantified and categorized by base substitution (x axis).
(D) The DT40 genome was divided into deciles by timing of replication from early (left) to late (right). Mutated cytidines in a TCN context are shown, where N is any base.
(E) Clustered mutations by the timing of replication. Top: number of mutation clusters (omikli and kataegis) per replication decile. Bottom: number of kataegis events per replication decile.
(F) APOBEC3A mutates DNA stem loops. Cytidine mutations in DT40-A3A genomes were compared with 38,325 control positions that represent the same trinucleotide spectrum. The ratio of APOBEC3A-induced mutations and control positions found in putative stem loops is categorized based on stem strength. The ratio was normalized to 1 in the “no stem loop” column (<3 bases in the stem).
(G) The difference between APOBEC3A and random stem loop mutation frequency is shown as a heatmap. The fraction of APOBEC3A mutations in each type ofstem loop (based on stem strength and loop length) relative to that of the control positions shows that the putative loops mutated by APOBEC3A in the DT40 genome have a stem strength of 5–25 and a loop length of 3–5.
(H) Sequence preference of APOBEC3A reflected in 3 nucleotide stem-loop mutations. TpC sites located entirely within the loop are mutated preferentially compared with loops where T is part of the stem structure.
(I) Bisulfite sequencing revealed the frequency with which each cytidine base within a CpG dinucleotide context in the DT40 genome was methylated (x axis). The proportion of cytidines mutated by APOBEC3A (red) and all other cytidines (gray) are indicated with respect to the likelihood that each cytidine base is methylated.