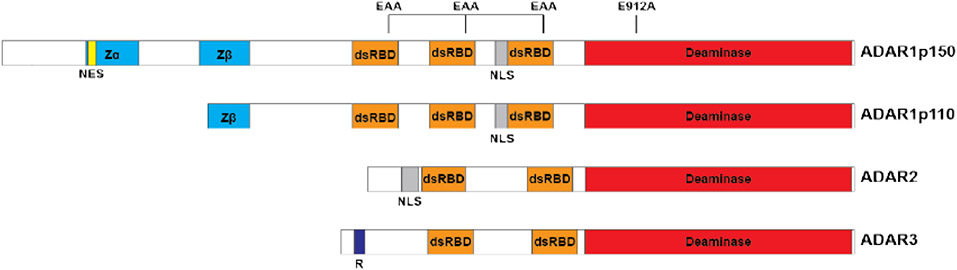
Figure 1. Schematic of the domain structure of the family of human ADARs.
ADAR1 is composed of two isoforms, p150 and p110, which share identical sequence with the exception of additional N-terminal sequence for p150, including an additional Zα domain and the NES which allows for both cytoplasmic and nuclear localization. Both contain three dsRBDs that mediate dsRNA binding and homodimerization, both of which are critical for enzymatic activity. Mutation of the KKxxK dsRBD motif to EAxxA abolishes dsRNA binding and thus editing activity. The E912A point mutation in ADAR1p150 (E617A for p110, E396A for ADAR2) disrupts catalytic deaminase activity. ADAR3 is thought to be catalytically inactive, and contains a unique arginine-rich domain that confers ability to bind single-stranded RNA. Abbreviations: NES, nuclear export signal; dsRBD, dsRNA binding domain; NLS, nuclear localization signal; R, arginine-rich domain; Zα/β, Z-DNA/RNA binding domains.