Figure 4. MH[sim] may interfere with Top2 processing of 359bp entanglements.
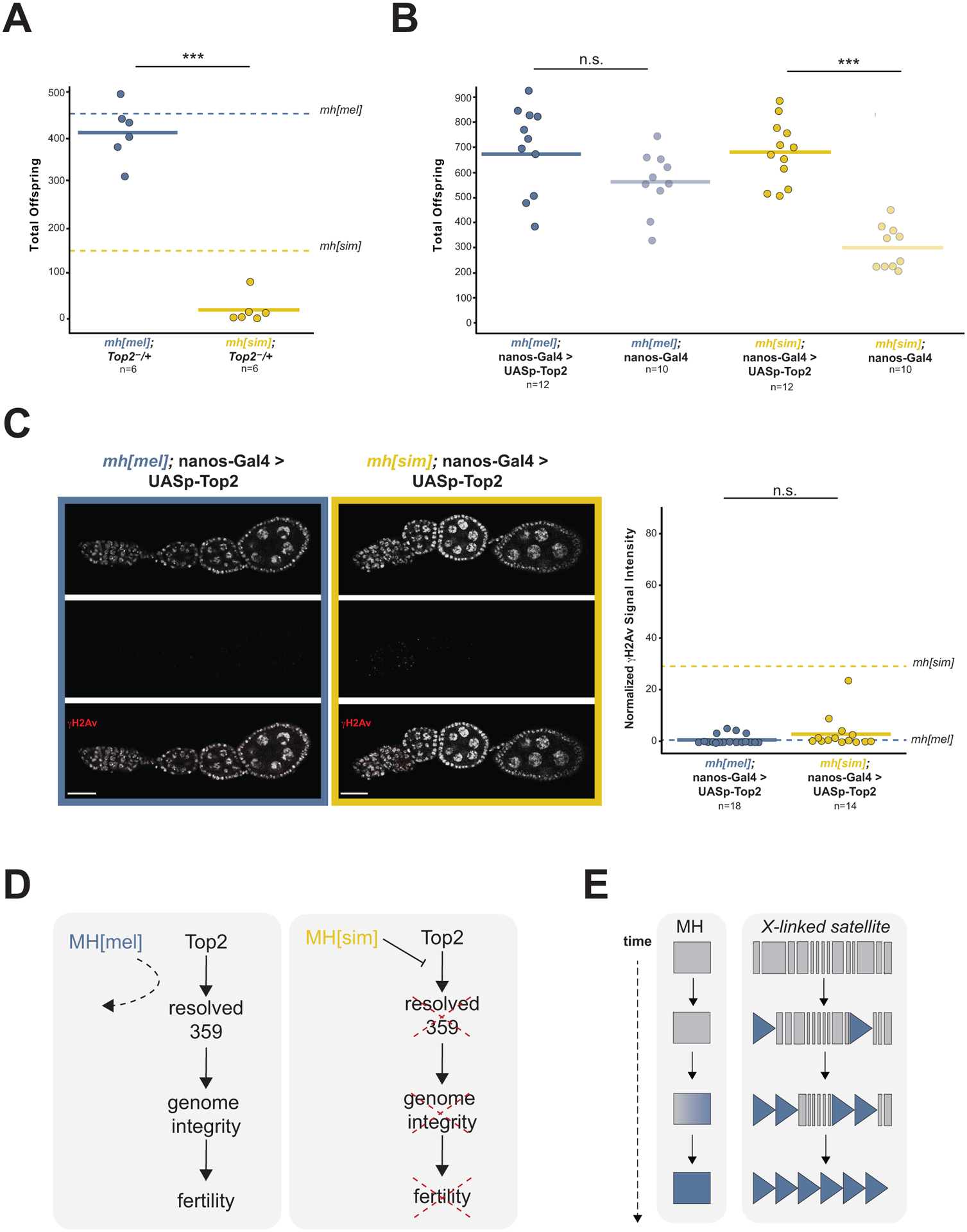
(A) Progeny counts from mh[mel]; Top2−/+ and mh[sim]; Top2−/+ females crossed to wildtype (w1118) males. Dotted lines correspond to mh[mel] and mh[sim] averages reported in Figure 1C. (B) Progeny counts from nos-Gal4-VP16 (female germline GAL4) driven mh[mel]; UASp-Top2 or mh[sim]; UASp-Top2 females crossed to wildtype (w1118) males. (C) γH2Av signal from ovaries of nos-Gal4-VP16 driven mh[mel]; UASp-Top2 or mh[sim]; UASp-Top2 females and quantification of normalized fluorescent signal intensity. Dotted lines correspond to mh[mel] and mh[sim] averages reported in Figure 2D. (D) Model of MH[sim] interference with Top2 processing of 359bp entanglements. These entanglements threaten genome integrity and ultimately, fertility. MH[mel], in contrast, has no measurable function in the ovaries, suggesting that it avoids interfering with 359bp processing by Top2. (E) Model of MH evolution tracking 359bp satellite proliferation. (t-test: “***” = p < 0.001, “n.s.” p > 0.05, scale bar = 25μm)
