Figure 5.
ILC depletion in Rag2−/− mice increases the severity of cystitis
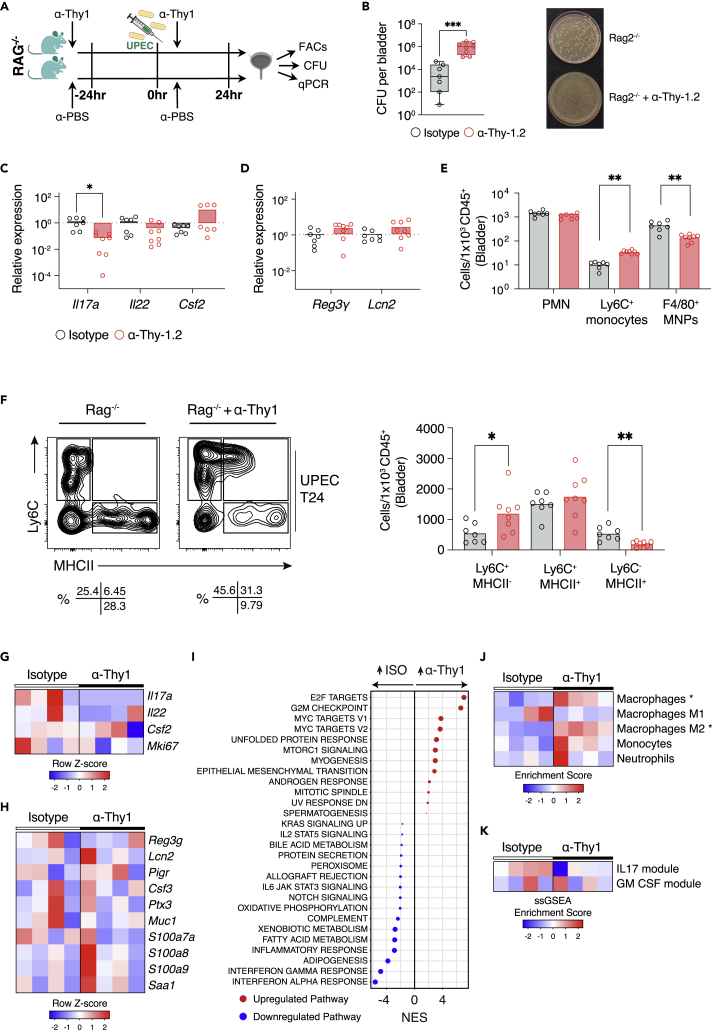
(A) Schematic of experimental design.
(B) Colony-forming units per bladder 24 h after infection with UTI89 in Rag2−/− + isotype (gray) and Rag2−/− + anti-Thy1 (red) mice (left panel) (Table S1) and corresponding image of bacterial growth on agar plates; 1:400 dilution (right panel). N = 7-8 mice per group.
(C and D) Corresponding qPCR of Th17 cytokines (C) and selected AMPs (D) in Rag2−/− + isotype (gray) and Rag2−/− + anti-Thy1 (red) bladders 24 h post infection (n = 6 per group). Results relative to Rag2−/− bladders. Data are representative of three independent experiments.
(E) Quantification of absolute cell counts in Rag2−/− + isotype (gray) and Rag2−/− + anti-Thy1 (red) bladders 24 h post infection (n = 7-8 per group) for the indicated subsets.
(F) Bladder “monocyte waterfall” subset quantification by flow cytometry 24 h post infection with UTI89 in Rag2−/− + isotype and Rag2−/− + anti-Thy1 bladders (n = 7-8 per group). Flow plots of CD45+Ly6G-CD11b+CX3CR1+ waterfall subsets (left) and quantification of absolute cell counts for the indicated subsets (right) are shown.
(G) Heatmap of Th17 cytokines from RNA sequencing of bladders infected with UTI89 in Rag2−/− + isotype (n = 4) and Rag2−/− + anti-Thy1 (n = 4) female mice 24 h after challenge. Data represent four biological replicates per group (isotype and anti-Thy1).
(H) Heatmap of selected IL22-dependent AMPs from (G).
(I) Gene set enrichment analysis of the differential expression from (G) against hallmarks pathways. Only significant pathways (FDR q value < 0.05) are plotted. Red dots indicate positive enrichment and blue negative, the size of the dot is inversely correlated with the FDR q value and the position indicates the normalized enrichment score (NES).
(J) Heatmap of cellular deconvolution of data in (G) using xCell (https://xcell.ucsf.edu). Scaled enrichment score is plotted (blue-red) with greatest enrichment in red.
(K) Heatmap of scaled enrichment scores from single-sample gene set enrichment analysis (ssGSEA, https://www.genepattern.org/modules/docs/ssGSEAProjection/4) of data in (G) for IL17a and GM-CSF signatures (up-regulated genes, p < 0.05, LFC>1.5). IL17a and GM-CSF signatures are derived from GEO: GSE20087 (Zhang et al, 2010) and GEO: GSE95404 (Zhang et al, 2010), respectively. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by Mann-Whitney test (B, D-E), one-way ANOVA with Dunn’s multiple comparisons test (C, F) and two-way ANOVA with Šídák’s multiple comparisons test (H, J, and K). All bladders used were from female mice unless otherwise stated.