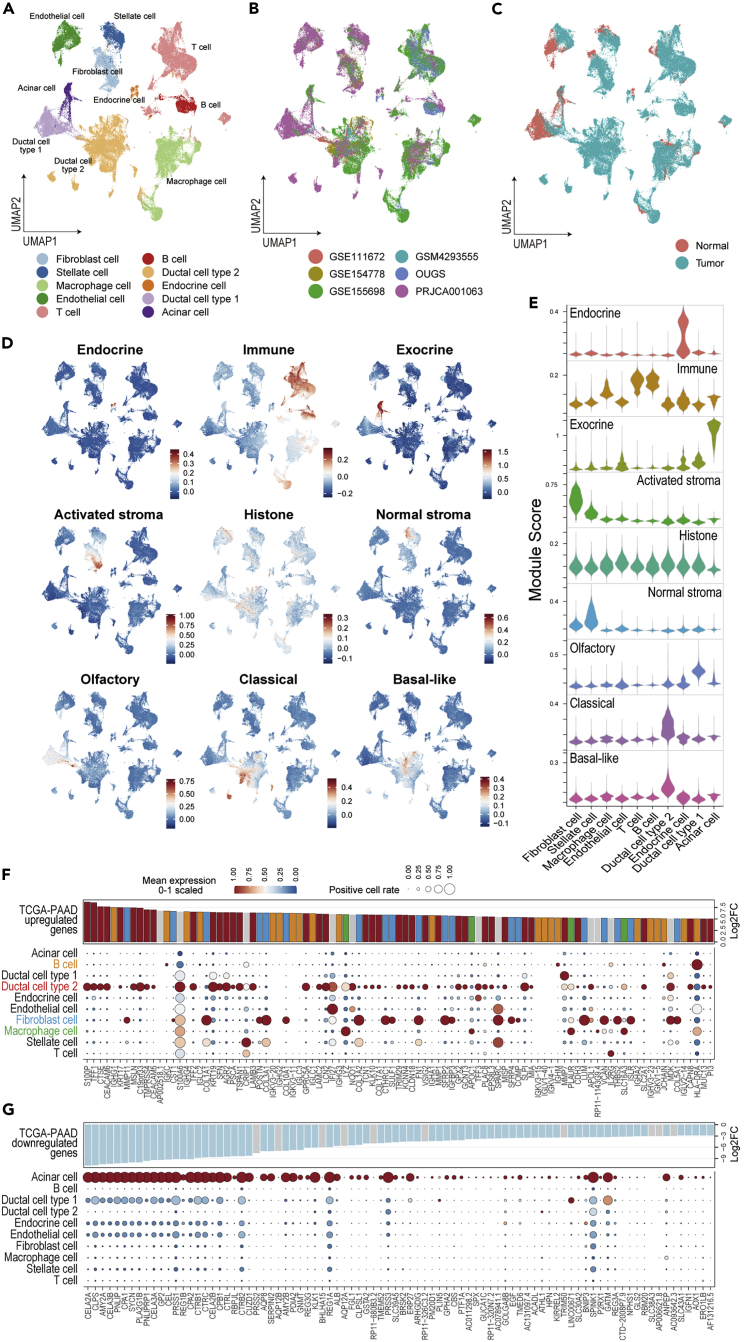
Figure 1.
Integration of the six scRNAseq datasets and reinterpretation of altered gene expression in bulk transcriptomics at the cellular level
(A–C) The integrated data are summarized in the UMAP and color-coded according to cell type (A), dataset (B), and sample type (C).
(D and E) The module scores calculated using the signatures defined in the NMF method on TCGA-PDAC are plotted on UMAP (D) and their violin plot (E).
(F and G) Bubble plot showing the cell origin of the driving genes that were highly upregulated (F) and downregulated (G) in TCGA-PAAD, with cell types indicated in the rows and genes in the columns.
The size of each bubble represents the rate of cells expressing the gene, and the color represents the scaled average expression in their cell type cluster. The bar plot shows the log2 fold change in the TCGA-PAAD sample vs. the control pancreas obtained from GEPIA2.