Figure 1.
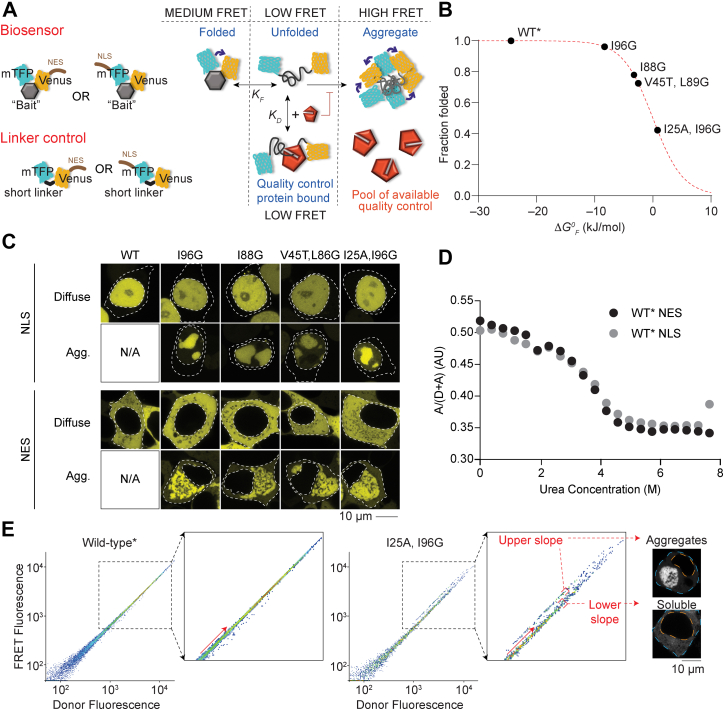
Targeting the barnase biosensor into the cytosol and nucleus.A, schematic of how the biosensor works (diagram adapted from Ref. (5)). The barnase protein is used as the “bait” for chaperones and flanked with fluorescence proteins for FRET measurements. A nuclear localization sequence (NLS) or nuclear export sequence (NES) is appended to the construct. B, shown is the relationship between mutations in barnase, the effect on standard free energy of folding (ΔG°F at 20 °C), and predicted fraction folded for the various biosensor variants used in the study. WT barnase is marked with ∗ to denote it contains the catalytic inactivation mutation H102A. This mutation is present in all constructs in the study. C, confocal images of HEK293T cells transiently transfected with either nuclear- or cytosol-targeting biosensor variants. The nucleus and cell edges are indicated by dashed lines. The localization of the biosensor is shown by Venus fluorescent protein fluorescence (yellow). Representative cells with only diffuse or aggregated proteins are shown. D, urea denaturation curves of WT∗ barnase biosensor variants as measured in cell lysates by FRET. E, flow cytometry strategy for monitoring foldedness and aggregation. Here, the donor and acceptor fluorescence of cells were measured by channels (FRET and donor fluorescence were gated by the PE (575/25) and V500 (525/50) filters, respectively, with the 405 nm laser). The inset highlights the changes that arise for cells bifurcated into “upper” and “lower” slope populations (division shown with red arrow). Representative cells collected from gates corresponding to the upper and lower slope populations imaged by confocal microscopy (grayscale). The orange dashed line denotes the nucleus boundary and the cyan dashed line the cell boundary. HEK293T, human embryonic kidney 293T cell line.