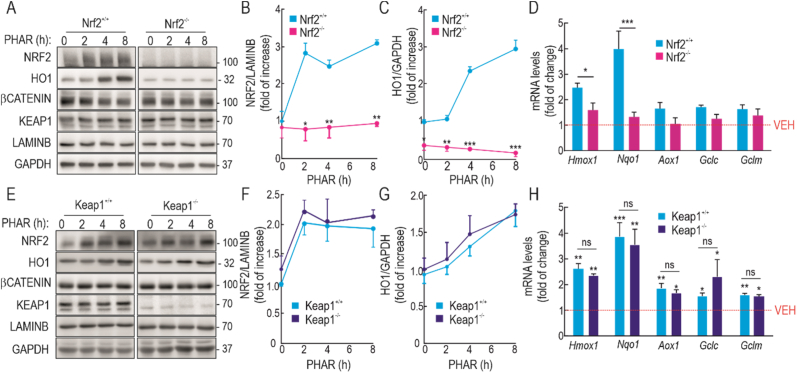
Fig. 3.
PHAR induces ARE-genes in an NRF2-dependent but KEAP1-independent manner. A, representative immunoblots of NRF2, HO1, β-CATENIN, KEAP1, LAMINB and GAPDH as a loading control from serum-depleted MEFs from wild type (Nrf2+/+) and NRF2-knockout (Nrf2−/−) mice subjected to 10 μM PHAR for the indicated times. B–C, densitometric quantification of NRF2 and HO1 protein levels from representative immunoblots from A, expressed as a ratio of LAMINB and GAPDH, respectively. Data are mean ± S.D. (n = 3). *p < 0,5; **p < 0,01; ***p < 0.001 vs vehicle of Nrf2+/+ according to a two-way ANOVA test. D, mRNA levels of several ARE-genes after 8 h of incubation with 10 μM PHAR were determined by qRT-PCR and normalized by the geometric mean of Gapdh, Tbp, and Actb levels. Data are mean ± S.D. (n = 4). *p < 0,5; ***p < 0.001 vs Nrf2+/+ according to a two-way ANOVA test. E representative immunoblots of NRF2, HO1, β-CATENIN, KEAP1, LAMINB, and GAPDH as a loading control from serum-depleted MEFs from wild type (Keap1+/+) and Keap1-knockout (Keap1−/−) mice subjected to 10 μM PHAR for the indicated times. F-G, densitometric quantification of NRF2 and HO1 protein levels from representative immunoblots from E, expressed as a ratio of LAMINB, and GAPDH, respectively. Data are mean ± S.D. (n = 3). H, mRNA levels of ARE-genes after 8 h of incubation with 10 μM PHAR were determined by qRT-PCR and normalized by the geometric mean of Gapdh, Tbp and Actb levels. Data are mean ± S.D. (n = 4). *p < 0.05; **p < 0.01; ***p < 0.001 vs Keap1+/+ according to a two-way ANOVA test; ns means non-significant differences between Keap1+/+ and Keap1−/−.