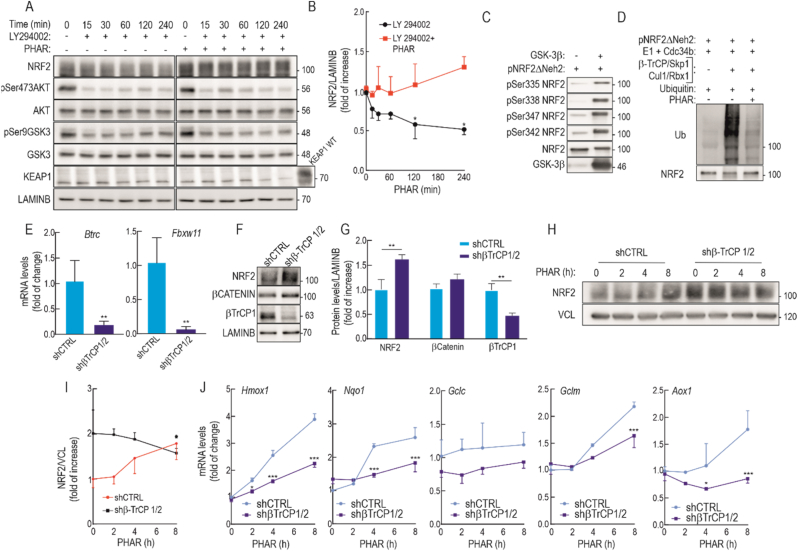
Fig. 4.
PHAR increases NRF2 protein levels in a β-TrCP dependent-manner. Serum-depleted Keap1−/− MEFs were subjected to the vehicle (0.1% DMSO) or 10 μM of PHAR for 30 min. Then, cells were treated with 20 μM LY294002 for the indicated times. A, representative immunoblots of NRF2, pSer473AKT, AKT, pSer9GSK3β, GSK3, KEAP1, and LAMINB as a loading control. B, densitometric quantification of NRF2 protein levels from representative immunoblots from A, expressed as a ratio of GAPDH. Data are mean ± S.D. (n = 3). *p < 0.05 vs LY2940002 at point 0, according to a two-way ANOVA test. C, In vitro kinase assay of NRF2 by GSK-3β. Recombinant pNRF2ΔNeh2, lacking the KEAP1 binding domain Neh2 [89], was submitted to an in vitro kinase assay in the absence or presence of recombinant GSK-3β. Representative immunoblots of pSer335NRF2, pSer338NRF2, pSer347NRF2, pSer342NRF2, NRF2, and GSK3β from in vitro kinase assay. D, In vitro ubiquitylation assay of NRF2 by the β-TrCP complex. Phospho-NRF2ΔNeh2 (20 ng) was incubated at 25 °C for 1 h with purified ubiquitin, E1/cdc34b, β-TrCP/Skp1, and Cul1/Rbx1 as indicated in the presence or absence of 1 μM PHAR. E, knockdown of Btrc (encoding β-TrCP1) and Fbxw11 (encoding β-TrCP2). Keap1−/− MEFs were transduced with control lentivirus encoding shCTRL or a combination of two lentiviruses encoding sh-Btrc and sh-Fbxw11. After 5 days, cells were serum-depleted for 16 h and then subjected to 10 μM PHAR for the indicated times. Transcript levels of Btrc and Fbxw11 were determined by qRT-PCR and normalized by the geometric mean of Gapdh, Tbp, and Actb. Data are mean ± S.D. (n = 4). *p < 0.05; **p < 0.01; ***p < 0.001 vs. shCTRL according to a Student's t-test. F, representative immunoblots of NRF2, β-CATENIN, β-TrCP1, and LAMINB as a loading control. G, densitometric quantification of NRF2, β-CATENIN, and β-TrCP1 protein levels from representative immunoblots from F, expressed as a ratio of LAMINB. Data are mean ± S.D. (n = 4). **p < 0.01 vs. shCTRL according to a Student's t-test. H, representative immunoblots of NRF2 and Vinculin (VCL) as a loading control, from control (shCTRL) and β-TrCP knocked-down (shβ-TrCP1/2) MEFs that were submitted to 10 μM PHAR for the indicated times. I, densitometric quantification of NRF2 protein levels from representative immunoblots from H, expressed as a ratio of NRF2/VCL. Data are mean ± S.D. (n = 4). *p < 0,05 vs. shCTRL at point 0 according to a one-way ANOVA test. J, expression of five NRF2-regulated genes in shCTRL vs. shβ-TrCP1/2 MEFs. Cells were submitted to 10 μM PHAR for the indicated times and transcript levels were determined by qRT-PCR and normalized by the geometric mean of Gapdh, Tbp, and Actb levels. Data are mean ± S.D. (n = 4). *p < 0.05; ***p < 0.001 vs. shCTRLat point 0 according to a two-way ANOVA test.