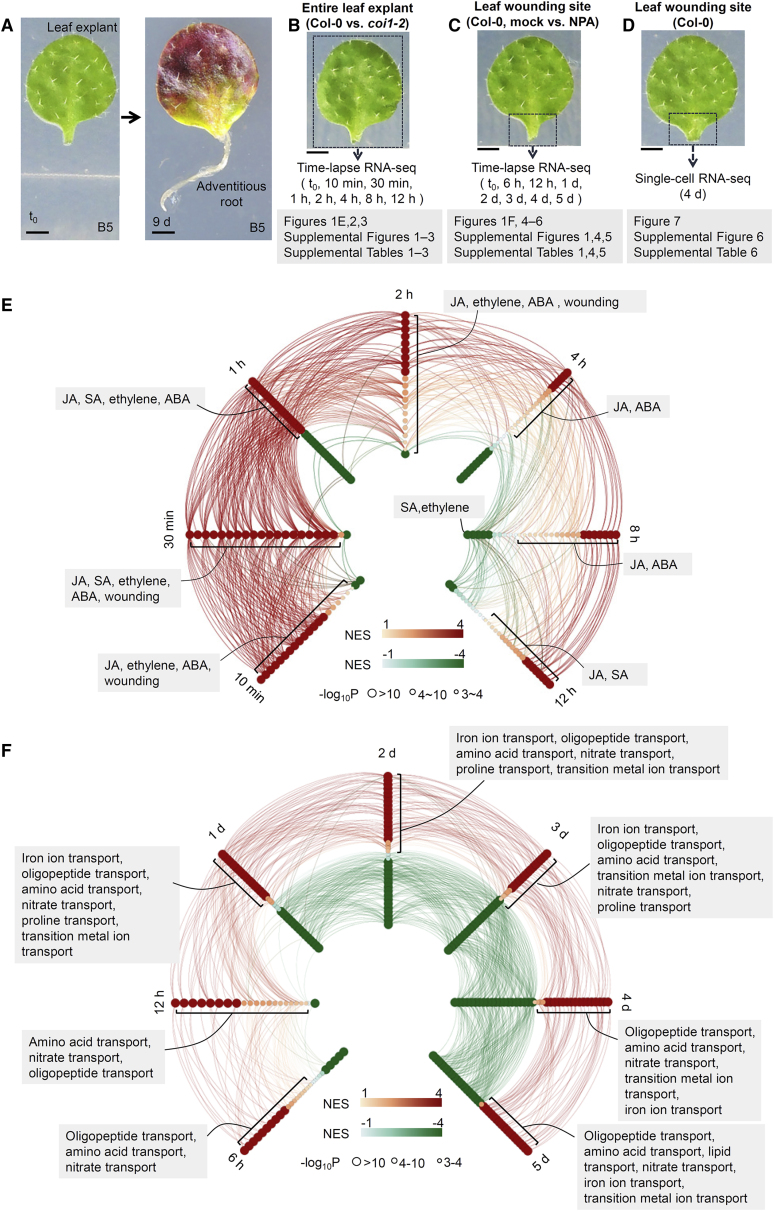
Figure 1.
Overview of RNA-seq experiments in this study.
(A) Adventitious rooting from Arabidopsis leaf explant.
(B) Time-lapse RNA-seq analysis of whole leaf explants (boxed region) of Col-0 and coi1-2 at t0, 10 min, 30 min, 1 h, 2 h, 4 h, 8 h, and 12 h after detachment. Two biological replicates were analyzed.
(C) Time-lapse RNA-seq analysis of wounded region of Col-0 leaf explant (boxed region) cultured on B5 medium without (mock) or with 5 μM NPA from t0, 6 h, 12 h, 1 d, 2 d, 3 d, 4 d, and 5 d after detachment. Because NPA was dissolved in dimethyl sulfoxide (DMSO), the mock control was treated with the same amount of DMSO in B5 medium. Two biological replicates were analyzed.
(D) Single-cell RNA-seq analysis of the wounded region of Col-0 leaf explant (boxed region) at 4 d after detachment. The data were from one biological replicate.
(E and F) Gene set enrichment analysis (GSEA) of GO terms enriched in up- or downregulated genes at each time point compared with t0. The time-lapse RNA-seq data for Col-0 (E, the entire detached leaf) or mock (F, the wounded region of detached leaves) were used for GSEA. Each dot represents the gene set of a GO term. Red dots represent gene sets enriched in upregulated genes (NES > 1), and green dots represent gene sets enriched in downregulated genes (NES < −1). GO terms with high gene overlap are connected by lines (enrichment of GO term pairwise comparison >5 and count of overlapped genes >20). GO terms related to JA, SA, ethylene, ABA, and wounding are listed in (E) as examples. GO terms related to transport are listed in (F) as examples. See Supplemental Table 1 for the full list of GO terms.
Scale bar, 1 mm in (A–D).