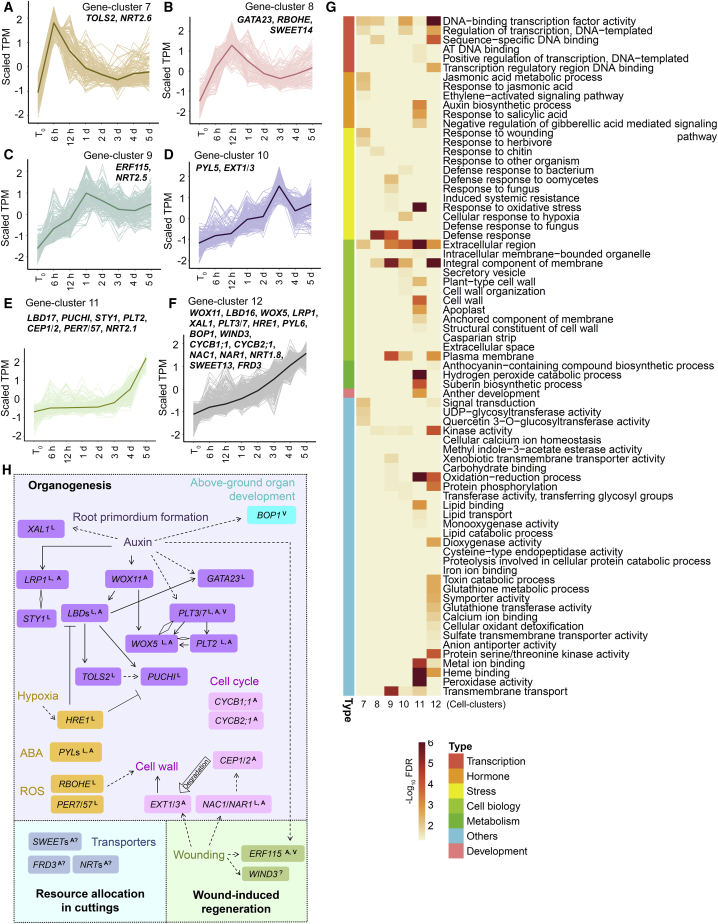
Figure 4.
Time-lapse RNA-seq analysis of candidate genes in adventitious root organogenesis.
(A–F) Candidate genes activated in the wounded region of Col-0 leaf explants (the mock without NPA treatment) cultured on B5 medium within 5 d after detachment, as determined from RNA-seq data (TPM < 2 at t0; log2[fold change] > 2 and FDR < 0.05 at any time point compared with that at t0). Candidate genes were grouped into six gene clusters (7–12) on the basis of changes in transcription patterns. Lines indicate average values.
(G) GO analysis of gene clusters 7 to 12. See Supplemental Table 4 for the full list of GO terms.
(H) Selected candidate genes identified in gene clusters 7 to 12. Genetic and molecular regulations are shown based on published data and this study. LValidated in lateral rooting by previous studies; Avalidated in adventitious rooting from hypocotyls and/or DNRR by previous studies; A?predicted or indicated in adventitious rooting in cuttings by previous studies; ?involved in regeneration but not yet validated in DNRR; Vvalidated in DNRR in this study. Rhombus indicates protein interactions.