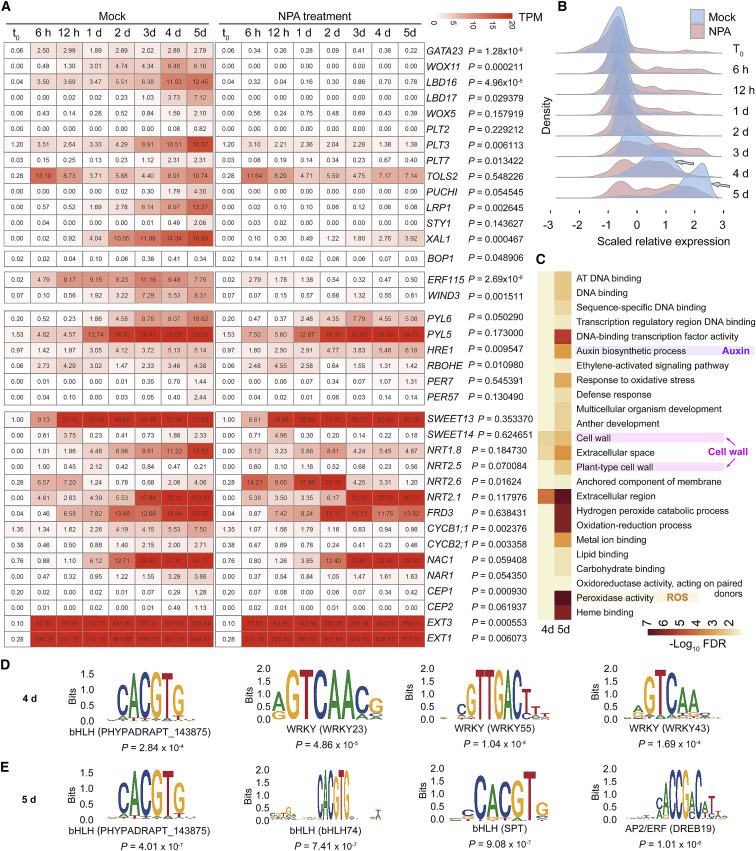
Figure 5.
Auxin control of gene activation.
(A) Transcript profiles of selected candidate genes in gene clusters 7 to 12 as determined from RNA-seq data from the wounded region of Col-0 leaf explants cultured on B5 medium (mock) or B5 medium with 5 μM NPA. Numbers in each box are TPM values. P values were calculated by performing a paired t-test between TPM values of Col-0 and coi1-2. One replicate was regarded as one sample. Note that HRE1 showed a higher expression level in the NPA treatment than in the mock control. Although the expression levels of WOX5 and PLT2 were relatively low in the RNA-seq data, the regulation of these two genes by auxin in DNRR was shown in previous studies (Hu and Xu, 2016; Bustillo-Avendaño et al., 2018).
(B) Distribution of genes with scaled relative expression at each time point from time-lapse RNA-seq data from the wounded regions of Col-0 leaf explants cultured on B5 medium (mock) or B5 medium with 5 μM NPA. Vertical lines indicate gene count density, and horizontal lines indicate transcript levels at each time point. We independently scaled the expression levels at each time point for mock and NPA treatment. Arrows show groups of genes that were highly upregulated in mock but not in NPA treatment at 4 and 5 d after leaf detachment.
(C) GO analysis of genes activated in mock but not in NPA treatment at 4 and 5 d after leaf detachment. Upregulation of these genes may be dependent on the auxin signaling pathway. See Supplemental Table 5 for the full list of GO terms.
(D and E) Analysis of promoter cis elements that may be targeted by the auxin pathway at 4 d (D) and 5 d (E) after leaf detachment. See Supplemental Table 5 for the full list of cis element analyses.