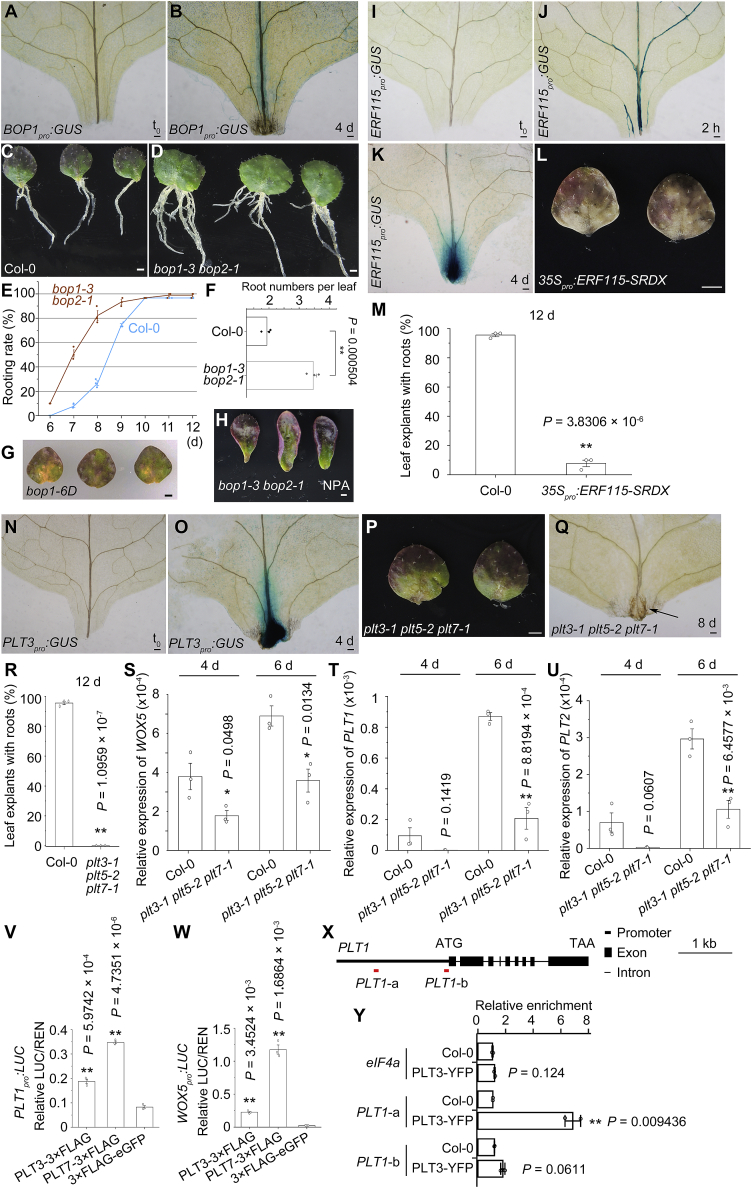
Figure 6.
BOPs, ERF115, and PLTs in DNRR.
(A and B) GUS staining of BOP1pro:GUS leaf explants at t0(A) and 4 d (B) cultured on B5 medium.
(C–F) Phenotype (C and D) and statistical (E and F) analysis of DNRR from wild-type Col-0 and bop1-3 bop2-1 leaf explants. Note that rooting rate (E) and root numbers per leaf explant at 15 d (C, D, and F) are higher in bop1-3 bop2-1 than in Col-0. Error bars show the SEM of three biological replicates (n = 30 in each replicate), and individual values are indicated by dots (E and F). ∗∗P < 0.01 in two-sided Student's t-test compared with Col-0 control (F).
(G) Phenotype analysis of bop1-6D. We tested 51 leaf explants from bop1-6D heterozygous or homozygous plants, and only two of them produced adventitious roots at 15 d.
(H) Phenotype analysis of bop1-3 bop2-1 leaf explants cultured on B5 medium with 5 μM NPA treatment. We tested 90 leaf explants, and none of them produced adventitious roots at 15 d.
(I–K) GUS staining of ERF115pro:GUS leaf explants at t0(I), 2 h (J), and 4 d (K) cultured on B5 medium.
(L) Phenotype analysis of DNRR from 35Spro:ERF115-SRDX leaf explants. See (C) for Col-0 control.
(M) Percentages of Col-0 and 35Spro:ERF115-SRDX leaf explants that regenerated roots by 12 d on B5 medium. Error bars show the SEM of three biological replicates (n = 30 in each replicate), and individual values are indicated by dots. ∗∗P < 0.01 in two-sided Student's t-test compared with Col-0 control.
(N and O) GUS staining of PLT3pro:GUS leaf explants at t0(N) and 4 d (O) cultured on B5 medium.
(P) Phenotype analysis of DNRR from plt3-1 plt5-2 plt7-1 leaf explants. See (C) for Col-0 control.
(Q) DIC observation of the wounded region of plt3-1 plt5-2 plt7-1 leaf explants. Arrow indicates the primordium-like structure. We observed 60 leaf explants from plt3-1 plt5-2 plt7-1, and all of them showed the primordium-like structure.
(R) Percentages of Col-0 and plt3-1 plt5-2 plt7-1 leaf explants that regenerated roots by 12 d on B5 medium. Error bars show the SEM of three biological replicates (n = 30 in each replicate), and individual values are indicated by dots. ∗∗P < 0.01 in two-sided Student's t-test compared with Col-0 control.
(S–U) qRT-PCR analysis of WOX5(S), PLT1(T), and PLT2(U) in the wounded region of leaf explants from Col-0 and plt3-1 plt5-2 plt7-1 at 4 and 6 d after leaf detachment. Error bars show the SEM of three biological replicates, and each biological replicate was analyzed with three technical replicates. ∗P < 0.05 and ∗∗P < 0.01 in two-sided Student's t-test compared with Col-0 control.
(V and W) Relative ratio of firefly LUC to Renilla luciferase (REN) activity in Arabidopsis protoplasts co-transformed with PLT1pro:LUC(V) or WOX5pro:LUC(W) with 35Spro:PLT3-3×FLAG or 35Spro:PLT7-3×FLAG. UBQ10pro:3×FLAG-eGFP served as the control. Error bars show the SEM of three biological replicates, and each biological replicate was analyzed with three technical replicates. ∗∗P < 0.01 in two-sided Student's t-test compared with control.
(X) Schematic of PLT1 gene structure. Red horizontal lines show positions of PCR fragments in ChIP analysis in (Y).
(Y) ChIP analysis showing relative enrichment of PLT3-YPF in the promoter of PLT1. eIF4a served as a negative control. Error bars show the SEM of two biological replicates, and each biological replicate was analyzed with three technical replicates. ∗∗P < 0.01 in two-sided Student's t-test compared with control.
Scale bars, 100 μm (A, B, I–K, N, O, and Q), 1 mm (C, D, L, G, H, and P).