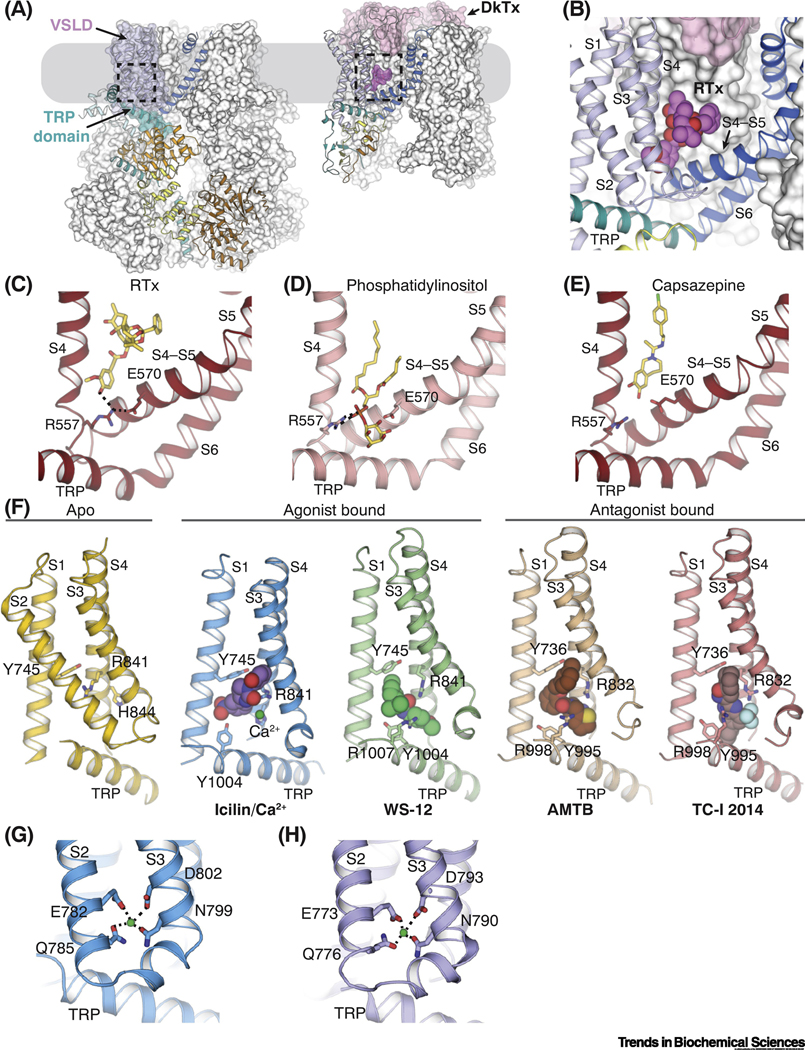
Figure 2. The novel binding site for agonists and antagonists in TRPM8.
(A) The locations of ligand binding in TRPM8 (left) and TRPV1 (right) embedded in the plasma membrane (gray box), which are highlighted by dashed lines. The ligand binding site in TRPM8 is located in the voltage-sensor-like domain cavity (VSLD cavity) formed by the VSLD and the TRP domain. The double-knot toxin (DkTx, pink surface representation) binds atop TRPV1 channel. For each channel, one protomer is shown as in Figure 1B and C, while the rest are shown in white surface representation.
(B) Close-up view of the binding site for resiniferatoxin (RTx) in TRPV1. Magenta spheres represent the RTx molecule.
(C-E) Comparison of the binding of vanilloid agonist RTx (C; PDB ID: 5IRX), phosphatidylinositol lipid (D; PDB ID: 5IRZ), and antagonist capsazepine (E; PDB ID: 5IS0) in TRPV1 channel. Dashed lines indicate interactions mediated by R557 in S4 and E570 in S4-S5 linker. Ligands are shown as sticks.
(F) Comparison of the VSLD cavity in the ligand-free TRPM8FA (yellow; PDB ID: 6BPQ), TRPM8FA-PI(4,5)P2/icilin/Ca2+ complex (blue; PDB ID: 6NR3), TRPM8FA-PI(4,5)P2/WS-12 complex (green; PBD ID: 6NR2), TRPM8PM-AMTB complex (wheat; PDB ID: 6O6R), and TRPM8PM-TCI-2014 complex (pink; PDB ID: 6O72). Key residues for ligand binding are shown in sticks. Spheres represent agonists, antagonists, and Ca2+ ion. In panels for ligand-bound structures, S2 is omitted for clarity.
(G-H) Ca2+ binding in the VSLD cavity of TRPM8FA-PI(4,5)P2/icilin/Ca2+ (G) and TRPM8PM-Ca2+ (H; PDB ID: 6O77) complex structures. Green spheres represent Ca2+ ions. Dashed lines indicate the ion coordination.