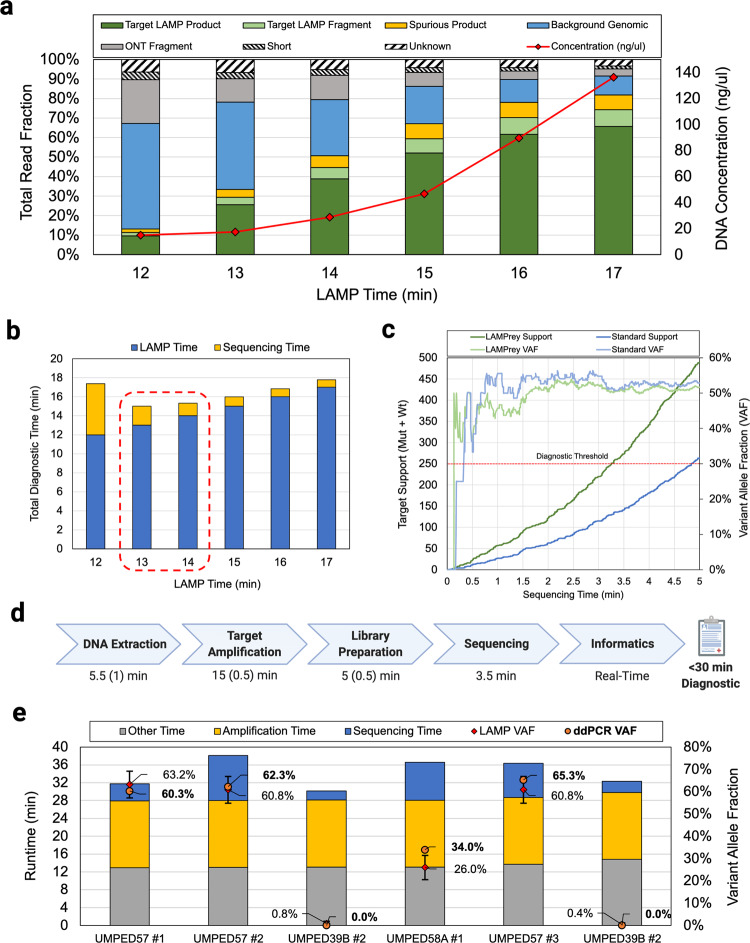
Fig. 5. LAMPrey ultra-rapid sequencing assay results.
a LAMPrey classification results from various amplification times and the final end-to-end demonstration (14 e2e) using DNA extracted from fresh tissue and the shortened LQE protocol. b Total sequencing time estimated from amplicon fraction at various LAMP time points. Our model predicted that 13–14 min of LAMP would lead to the fastest end-to-end diagnostic. We chose to evaluate 14 min to guard against under-amplification or other sequencer performance model misprediction error. c Diagnostic results of the 14-min end-to-end LAMP assay over time. A standard bioinformatics pipeline failed to leverage ~48% of target reads. LAMPrey can recover this information and leads to a diagnosis ~96 s sooner. d Approximate times for each diagnostic step with approximate manual overhead in parenthesis. Library preparation is notably faster than the 3-step PCR evaluation due to reduced adapter incubation time and ice quenches. Amplification time is lower due to our 14-minute LAMP protocol vs ~26-minute (28 cycle) PCR amplification protocol. e Results from barcoded protocol runs after prediction model refinement showing results over multiple patients (n = 3), samples (n = 6), and flow cells (n = 3). Replicates were performed with barcoded library preparation kit and without ice-plunges. An updated sequencer performance model estimated 15-minutes as the optimal amount of amplification. VAFs reported by ddPCR using the same DNA from each ultra-rapid run closely match those reported by LAMPrey (error bars computed using two-sided proportion confidence interval CL = 95%).