Figure 5.
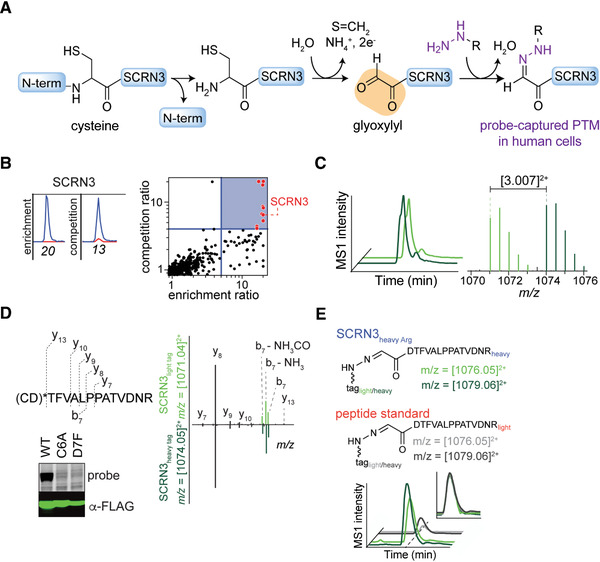
Sample data from RP‐ABPP experiments. Adapted from published work (Matthews et al., 2017). (A) Reaction scheme of probe captured glyoxylyl on SCRN3. (B) Extracted parent ion chromatograms and corresponding H/L ratios for tryptic peptides of the SCRN3 protein of probe quantified in enrichment and competition (left) and quadrant plot of average competition versus enrichment SILAC ratios from quantitative proteomics experiments (right). (C) Extracted MS1 ion chromatograms (left) and corresponding isotopic envelopes (right) for coeluting heavy‐ and light‐tagged peptides labeled by probe (in dark and light green, respectively). (D) Comparison of high‐resolution MS2 spectra generated from light‐ versus heavy‐tagged parent ions. The y‐ions resolve the modified site (*) to the N‐terminal cysteine and/or adjacent aspartate. Probe‐labeling and expression profiles of Cys6‐to‐Ala6 (C6A) and Asp7‐to‐Phe7 (D7F) mutant SCRN3 proteins compared to wild‐type (WT) SCRN3. (E) Heavy‐Arg/Lys‐labeled SCRN3‐transfected cells treated with probe, and then processed by isoTOP‐ABPP, furnishes an isotopically differentiated probe‐labeled SCRN3 peptide pair (light and dark green), which coelutes with a light‐amino‐acid‐labeled probe–glyoxylyl6‐Arg20 standard (also an isotopically differentiated peptide pair; light and dark gray). Inset chromatogram shows all four traces scaled to the same intensity (inset plot) to show coelution of endogenous and standard probe–glyoxylyl6‐Arg20 SCRN3 peptides.
