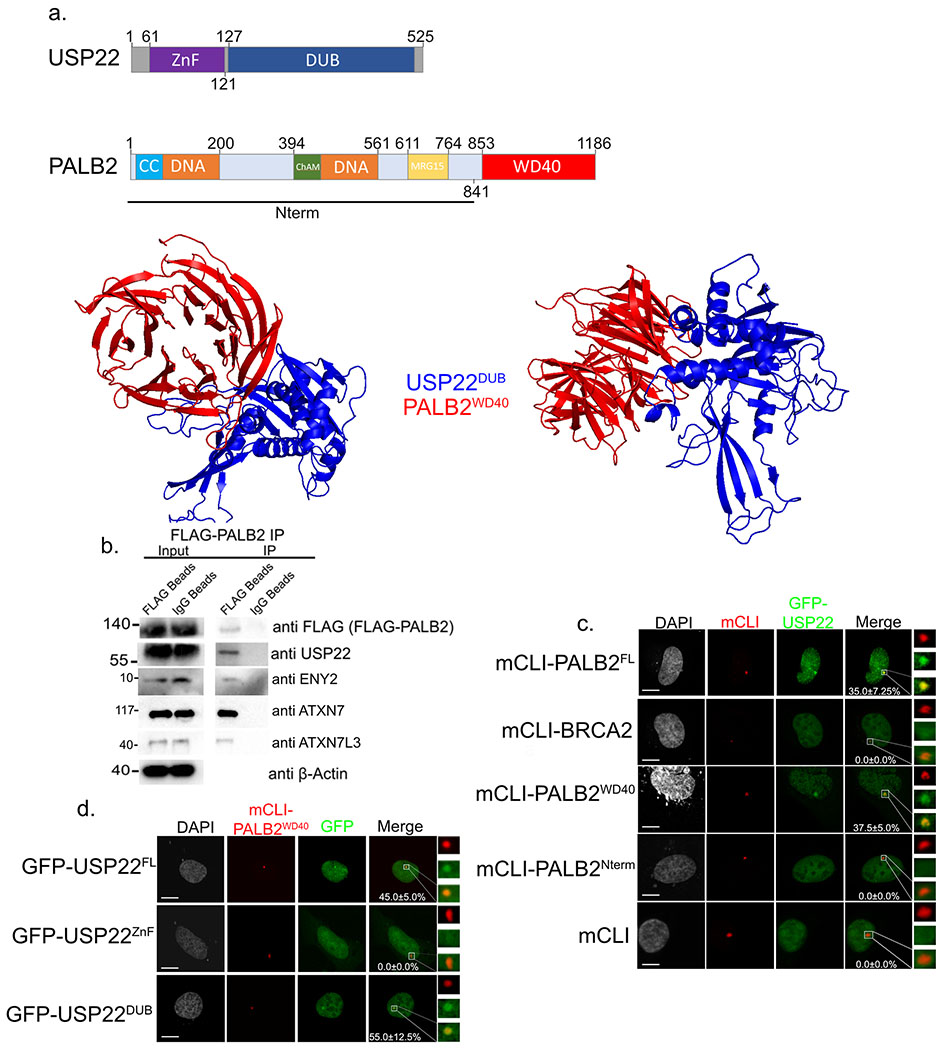
Figure 3:
USP22 DUB domain interacts with PALB2 WD40 domain. a. Cartoon of different domains of USP22 and PALB2 (upper). Computer model using ZDOCK predicting USP22 DUB (blue) domain binding to PALB2 WD40 domain (red). b. Immunoprecipitation of FLAG-PALB2 plasmid transiently transfected into H1299 cells. Western blot against FLAG (FLAG-PALB2), USP22, ENY2, ATXN7L3, ATXN7, and β-Actin as a loading control in the input. Numbers to the left of western blots indicate molecular weight. c. U2OSFOKI cells co-transfected with the indicated mCherry-LacI protein to tether it to 256X LacO array along with GFP-USP22. mCherry-LacI alone is used as a negative control. Percentage of cells with recruitment of GFP-USP22 to LacO array are indicated with +/− SD. White scale bars indicated 5um. 120 cells were counted for each condition for every experimental round. Experiment was done in triplicate. d. U2OSFOKI cells co-transfected with mCherry-LacI-PALB2WD40 protein to tether it to 256X LacO array along with indicated GFP-USP22 fragment. Percentage of cells with recruitment of GFP-USP22 to LacO array are indicated with +/− SD. White scale bars indicated 5um. 120 cells were counted for each condition for every experimental round. Experiment was done in triplicate. For all statistical analysis a student’s two-tailed t-test was used to establish p-value.