Figure 4.
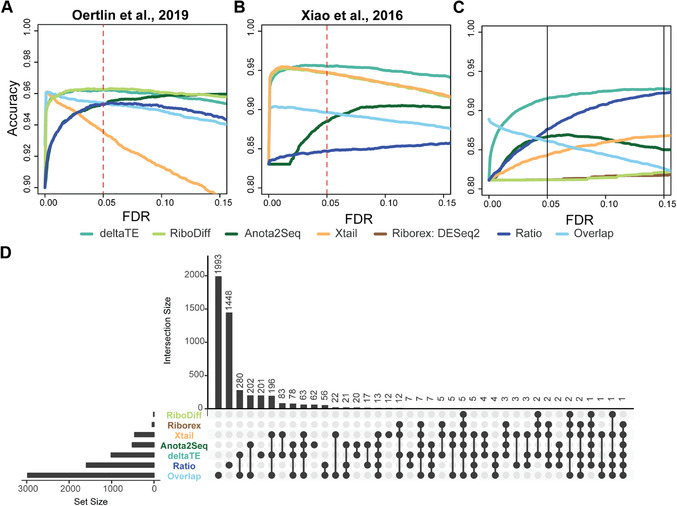
Benchmarking of published tools to detect differential translation efficiency genes (DTEGs). Simulation datasets (A) derived from Oertlin et al. (2019), (B) derived from Xiao et al. (2016), and (C) generated using the Polyester package to introduce batch effects were used. All three simulations show that ΔTE outperforms all other published methods. Comparisons are made using all the DTEGs as the true set. Since Anota2Seq has two different functions for obtaining exclusive and buffered genes, the results are combined prior to comparison. Riborex is omitted in simulated datasets without batch effects (A, B), since it is equivalent to the ΔTE approach in these cases. The ratio method is based on quantifying the ratio of DESeq2 fold changes for mRNA counts and RPF. The overlap method identifies DTEGs as genes which have either significantly changing mRNA counts or RPFs but not both. (D) Analysis on published data showed inability of previous tools to reliably identify DTEGs.
